Search Count: 844
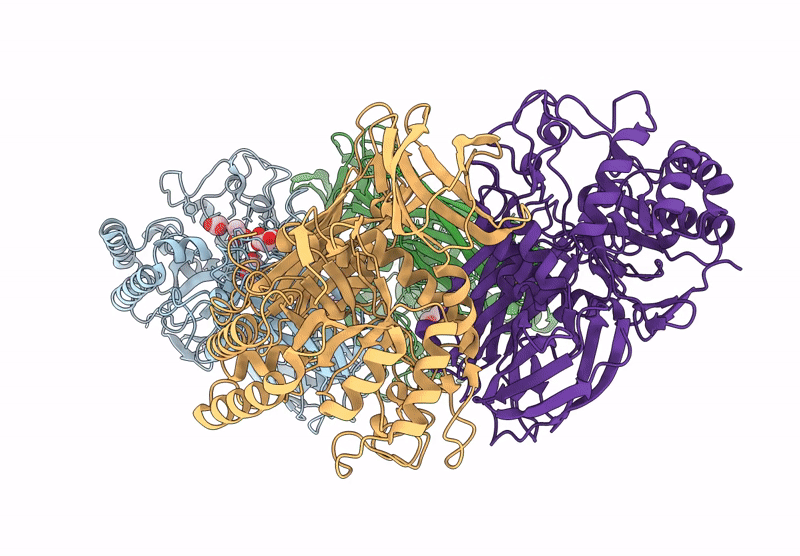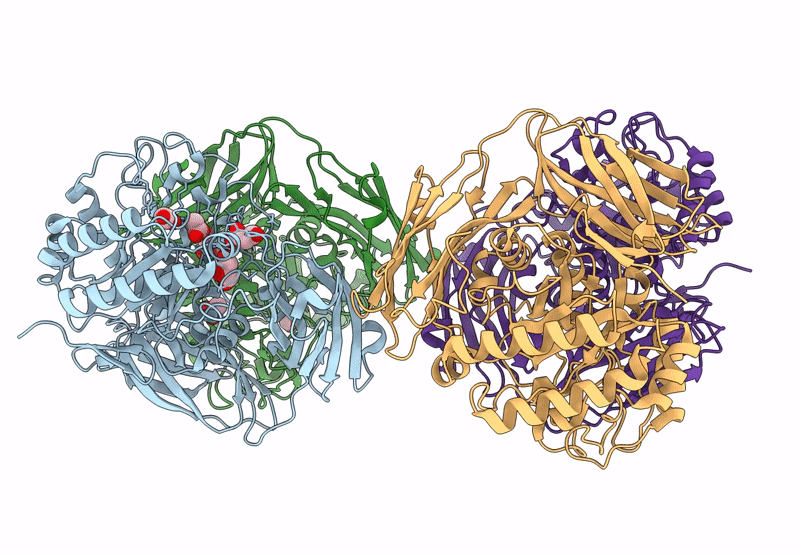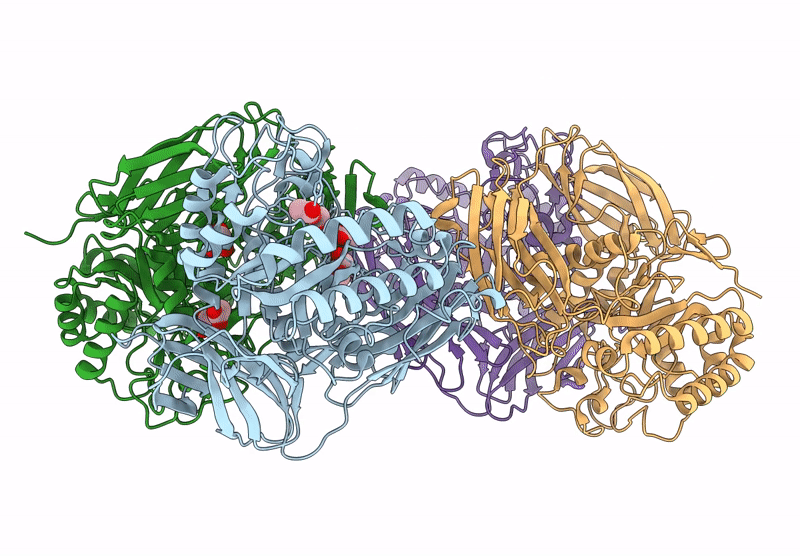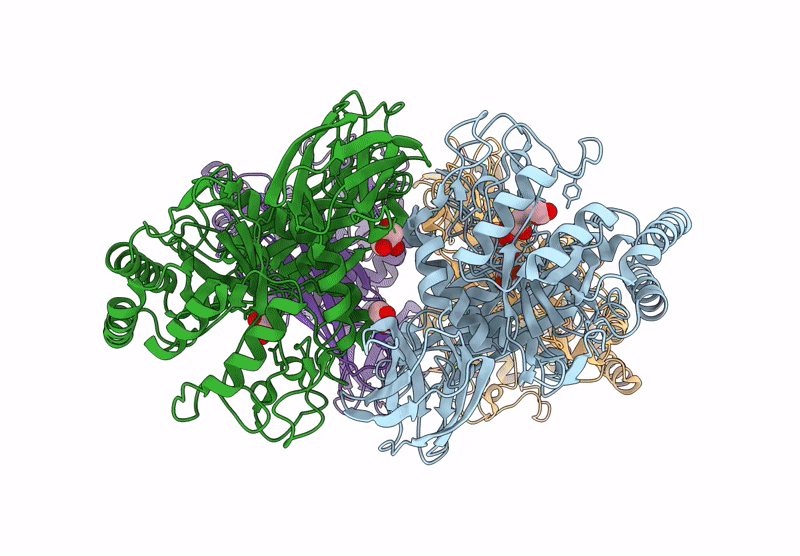 |
Glycoside Hydrolase Family 157 From Labilibaculum Antarcticum, Wild Type Semet Derivative (Lagh157)
Organism: Labilibaculum antarcticum
Method: X-RAY DIFFRACTION Release Date: 2025-07-16 Classification: HYDROLASE Ligands: ACT, GOL, MLA |
 |
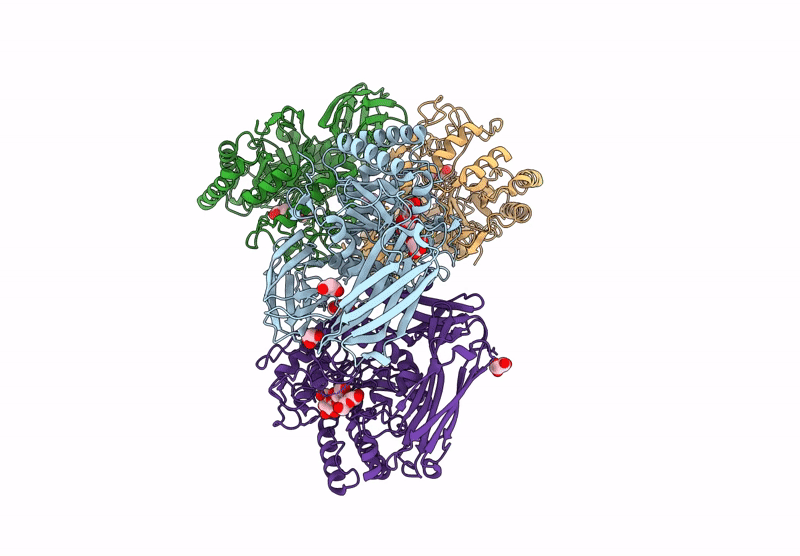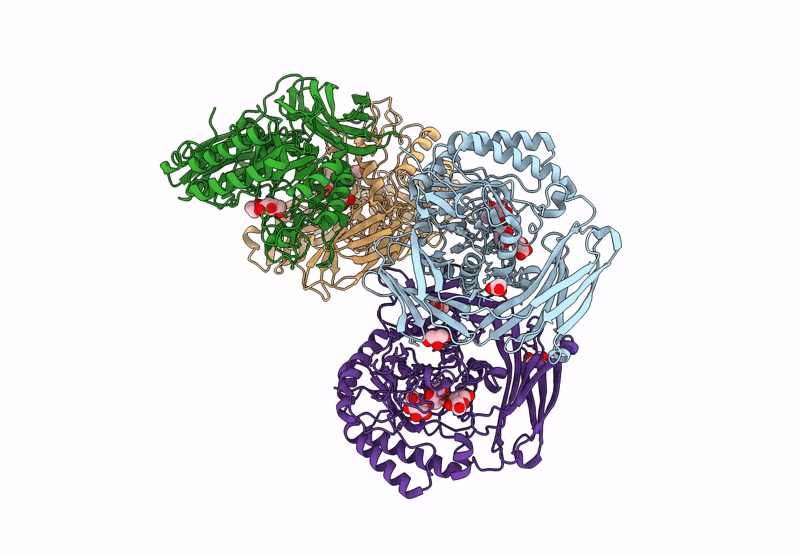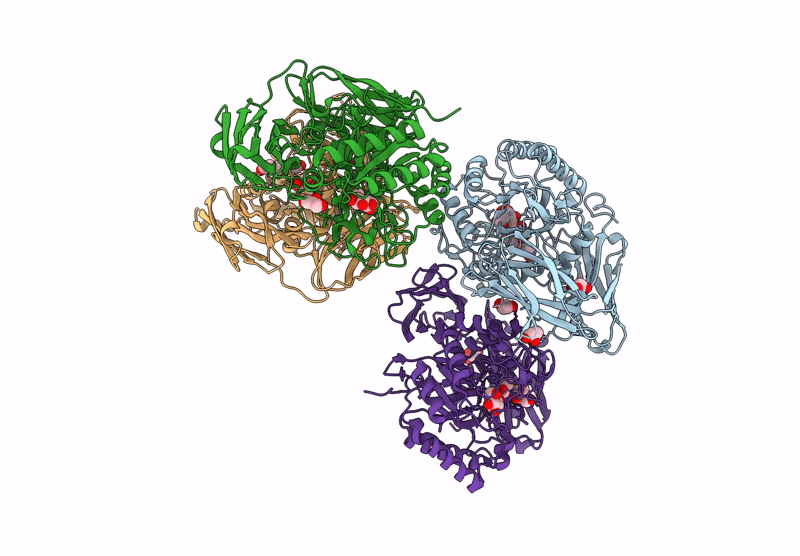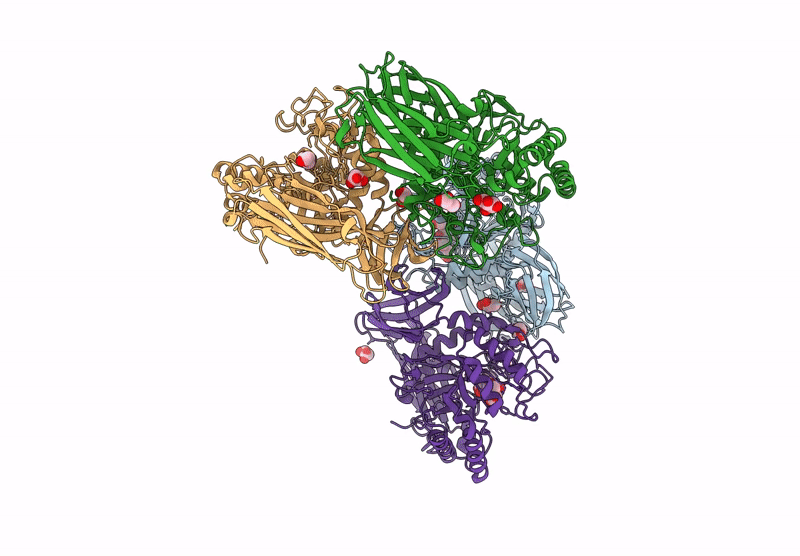
Glycoside Hydrolase Family 157 From Labilibaculum Antarcticum (Lagh157) E224A Mutant In Complex With Laminaritriose And Glucose
Organism: Labilibaculum antarcticum
Method: X-RAY DIFFRACTION Resolution:2.32 Å Release Date: 2025-05-21 Classification: HYDROLASE Ligands: EDO, BGC, GOL, PGE |
 |
Glycoside Hydrolase Family 157 From Labilibaculum Antarcticum (Lagh157) In Complex With Laminaribiose
Organism: Labilibaculum antarcticum
Method: X-RAY DIFFRACTION Resolution:2.71 Å Release Date: 2025-05-21 Classification: HYDROLASE Ligands: MLA, GOL, BGC, PG4 |
 |
Gh191 Family Alpha-Galactosaminidase From Environmental Sample (99.2% Identity To Myxococcus Fulvus Enzyme)
Organism: Environmental samples
Method: X-RAY DIFFRACTION Resolution:1.64 Å Release Date: 2025-04-23 Classification: HYDROLASE Ligands: SO4 |
 |
Organism: Fusarium solani
Method: X-RAY DIFFRACTION Resolution:1.61 Å Release Date: 2025-04-23 Classification: HYDROLASE Ligands: EDO, PEG |
 |
Organism: Thermotoga maritima msb8
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2025-04-09 Classification: HYDROLASE |
 |
Organism: Thermotoga maritima msb8
Method: X-RAY DIFFRACTION Resolution:2.60 Å Release Date: 2025-04-09 Classification: HYDROLASE |
 |
Organism: Terribacillus saccharophilus
Method: X-RAY DIFFRACTION Resolution:1.95 Å Release Date: 2025-03-12 Classification: HYDROLASE Ligands: ACT |
 |
Organism: Terribacillus saccharophilus
Method: X-RAY DIFFRACTION Resolution:1.51 Å Release Date: 2025-03-12 Classification: HYDROLASE Ligands: GC2, MPD |
 |
Organism: Lactiplantibacillus paraplantarum
Method: X-RAY DIFFRACTION Resolution:1.05 Å Release Date: 2025-03-12 Classification: HYDROLASE Ligands: GC2, EDO, PEG |
 |
Dispersin From Terribacillus Saccharophilus Dispts2 In Complex With Nag-Thiazoline
Organism: Terribacillus saccharophilus
Method: X-RAY DIFFRACTION Resolution:2.17 Å Release Date: 2025-03-12 Classification: HYDROLASE Ligands: NGT |
 |
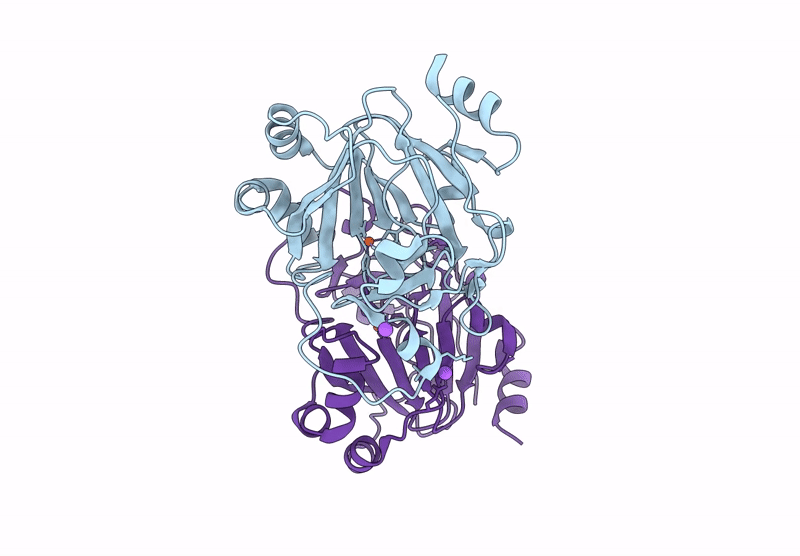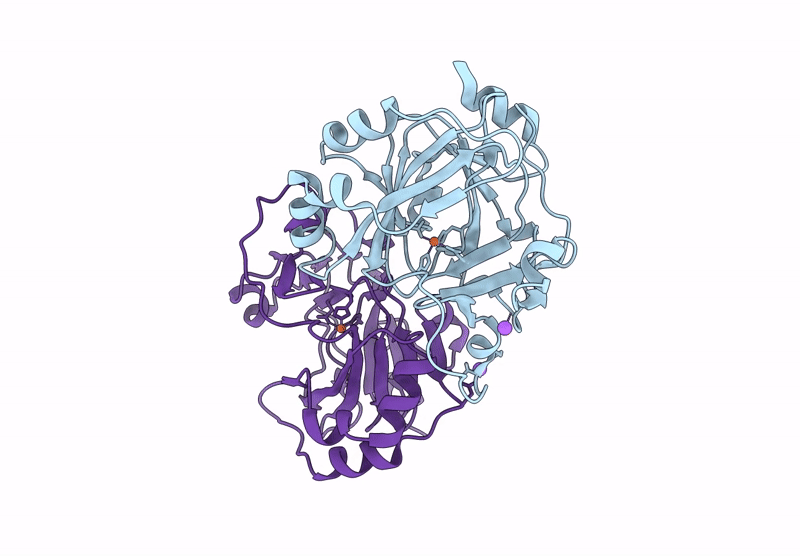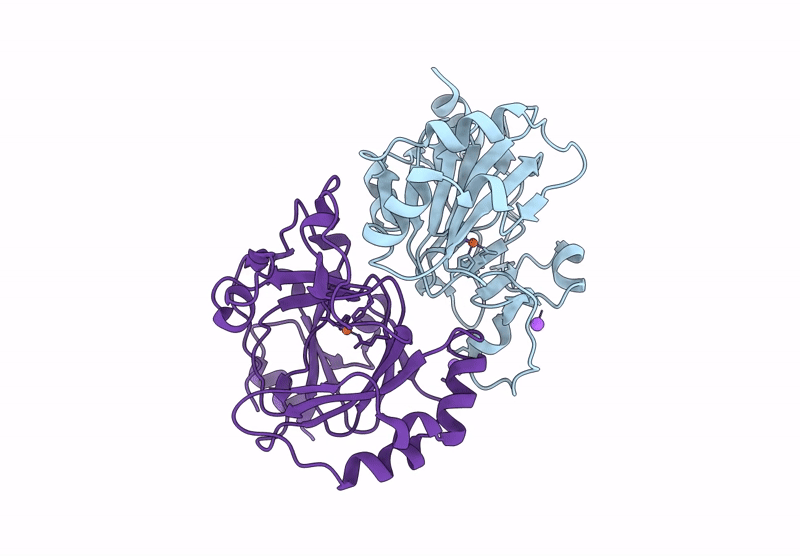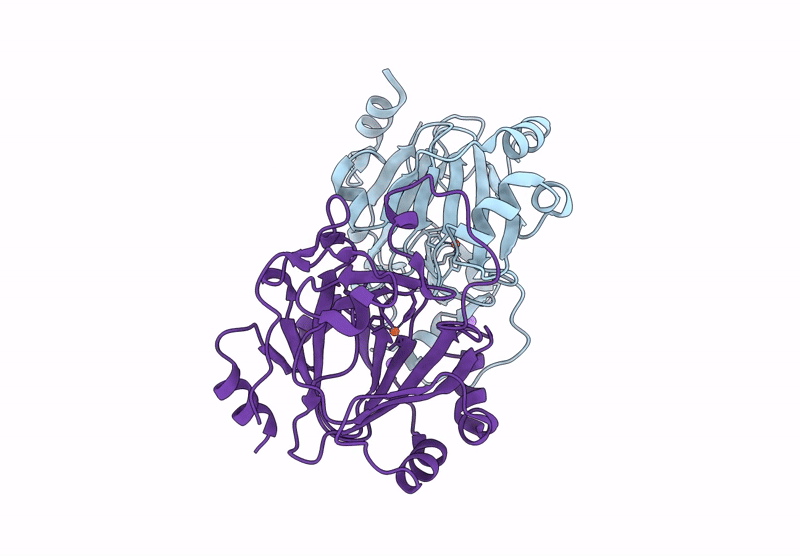
Organism: Pseudomonas aeruginosa
Method: X-RAY DIFFRACTION Resolution:1.55 Å Release Date: 2025-03-05 Classification: HYDROLASE Ligands: CL, NA, GOL |
 |
Organism: Pseudomonas aeruginosa
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2025-03-05 Classification: HYDROLASE Ligands: GOL, CL |
 |
Crystal Structure Of Domain-Of-Unknown-Function Duf4867 From Bacillus Megaterium
Organism: Priestia megaterium
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2025-02-26 Classification: ISOMERASE Ligands: FE, NA, CL |
 |
Crystal Structure Of Domain-Of-Unknown-Function Duf4867 From Bacillus Megaterium (Unmodelled Additional Ligand Density At Active Site)
Organism: Priestia megaterium
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2025-02-26 Classification: ISOMERASE Ligands: FE |
 |
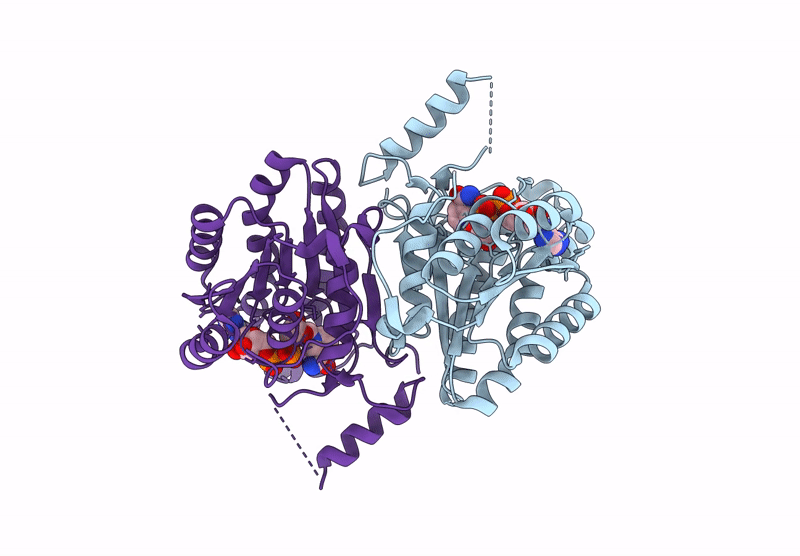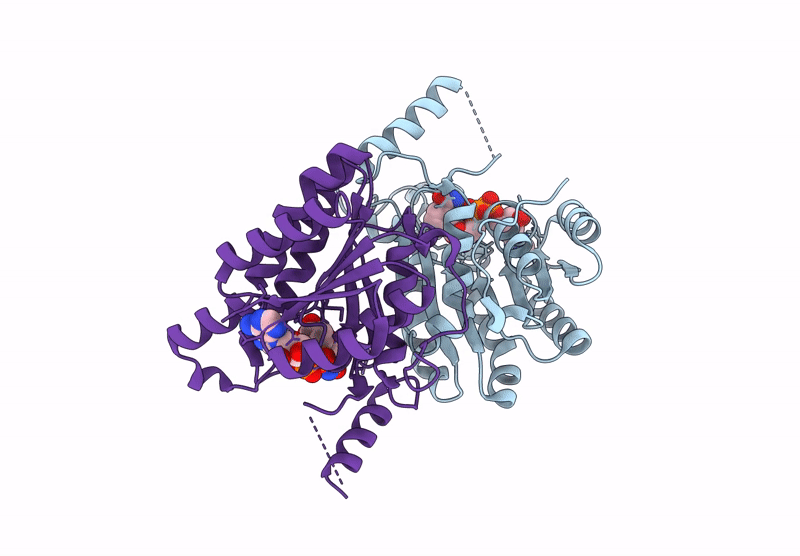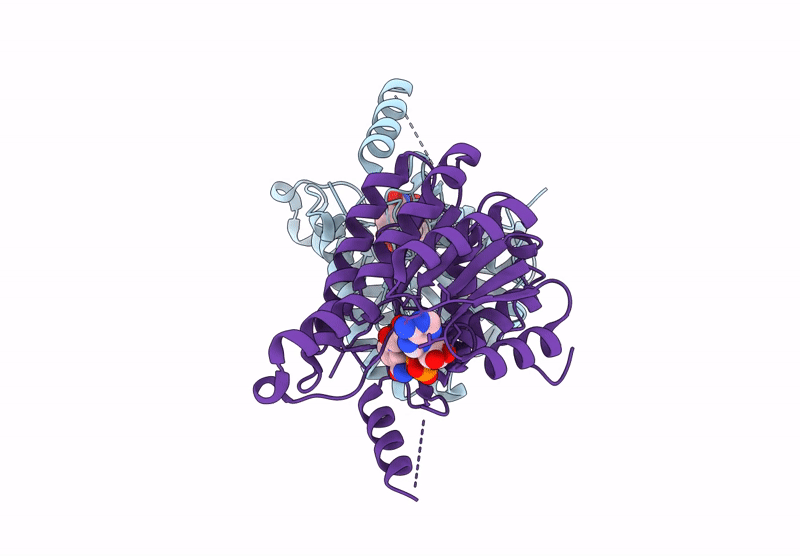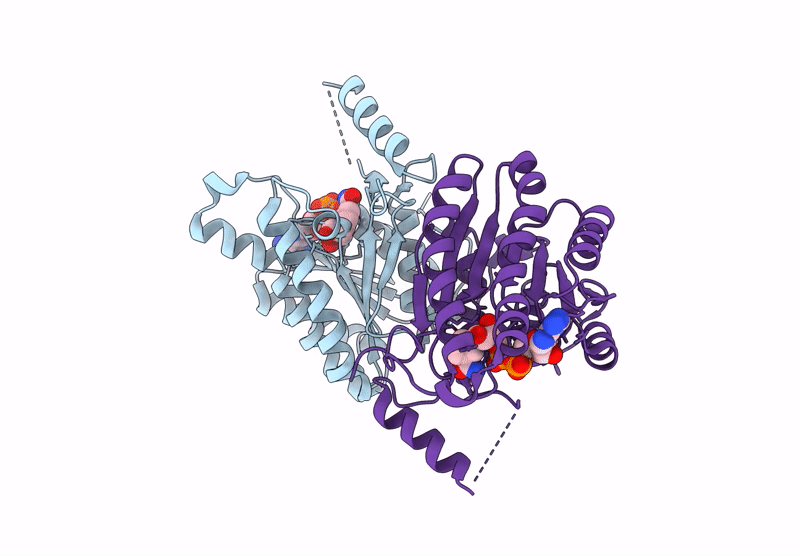
Crystal Structure Of Sulfoquinovose-1-Dehydrogenase From Pseudomonas Putida (Sulfo-Ed Pathway)
Organism: Pseudomonas putida
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2025-02-12 Classification: OXIDOREDUCTASE |
 |
Crystal Structure Of Sulfoquinovose-1-Dehydrogenase From Pseudomonas Putida In Complex With Nad+ (Sulfo-Ed Pathway)
Organism: Pseudomonas putida
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2025-02-12 Classification: OXIDOREDUCTASE Ligands: NAD |
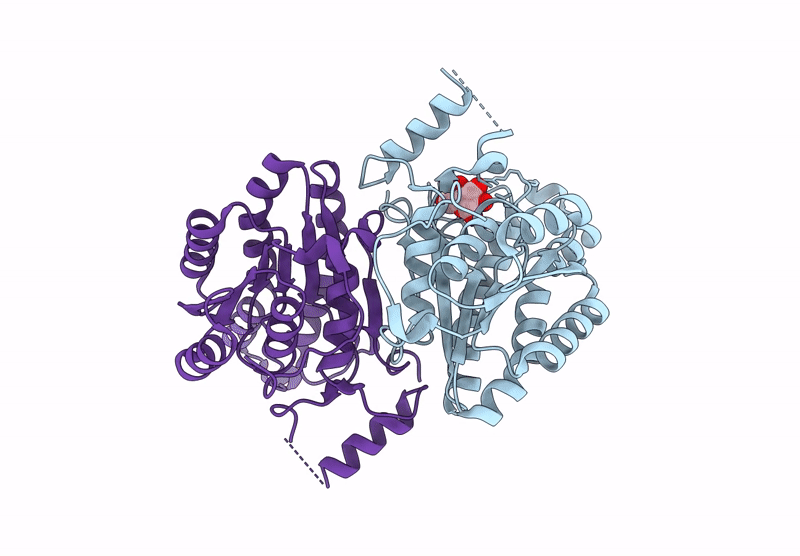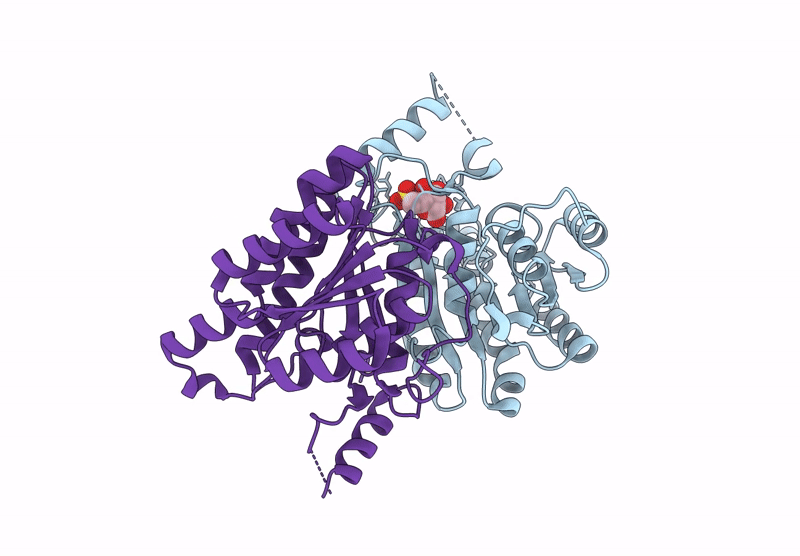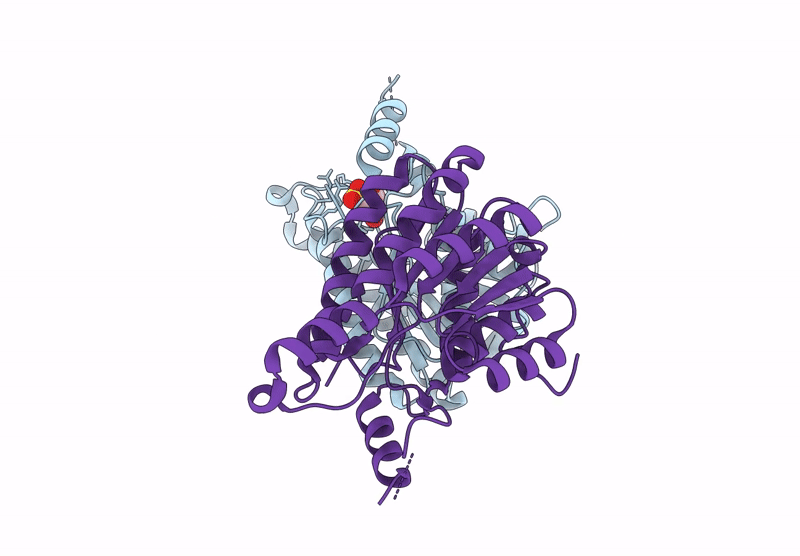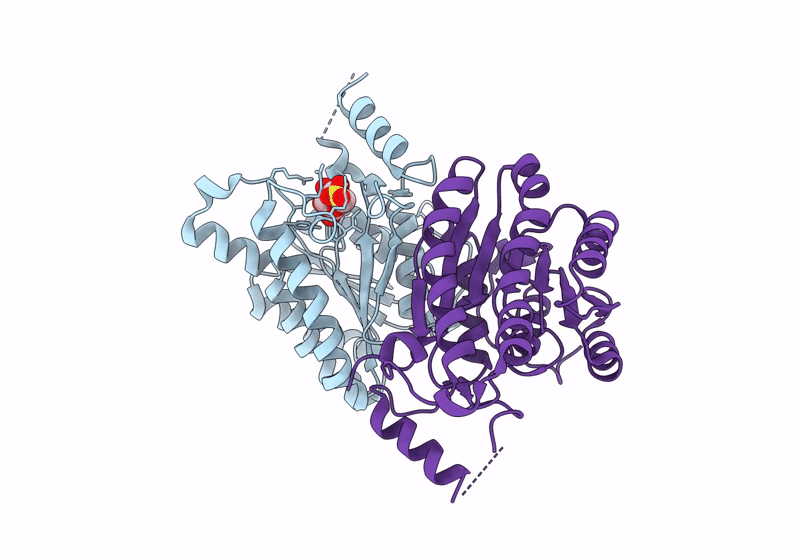 |
Crystal Structure Of Sulfoquinovose-1-Dehydrogenase From Pseudomonas Putida In Complex With Sulfoquinovose Substrate (Sulfo-Ed Pathway)
Organism: Pseudomonas putida
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2025-02-12 Classification: OXIDOREDUCTASE Ligands: YZT |
 |
Crystal Structure Of Susa Amylase From Bacteroides Thetaiotaomicron Covalently Bound To Alpha-1,6 Branched Pseudo-Trisaccharide Activity-Based Probe
Organism: Bacteroides thetaiotaomicron
Method: X-RAY DIFFRACTION Resolution:2.43 Å Release Date: 2024-12-11 Classification: HYDROLASE Ligands: PBW, OC9, IMD, CA |
 |
Crystal Structure Of Susg From Bacteroides Thetaiotaomicron Covalently Bound To Alpha-1,6 Branched Pseudo-Trisaccharide Activity-Based Probe
Organism: Bacteroides thetaiotaomicron
Method: X-RAY DIFFRACTION Resolution:2.65 Å Release Date: 2024-12-11 Classification: HYDROLASE Ligands: PBW, OC9, A1ILG, CA, IMD, ACT |

