Planned Maintenance: Some services may turn out to be unavailable from 15th January, 2026 to 16th January, 2026. We apologize for the inconvenience!
Planned Maintenance: Some services may turn out to be unavailable from 15th January, 2026 to 16th January, 2026. We apologize for the inconvenience!
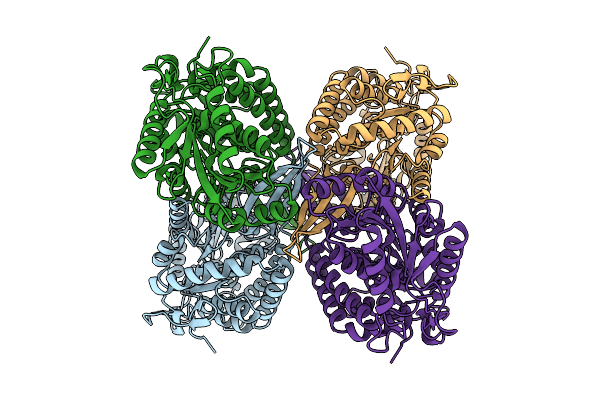 |
Cryoem Structure Of Aldehyde Dehydrogenase From Burkholderia Cenocepacia At 2.33A Resolution
Organism: Burkholderia cenocepacia
Method: ELECTRON MICROSCOPY Release Date: 2025-12-10 Classification: OXIDOREDUCTASE |
 |
Cryoem Structure Of Methylmalonic Acid Semialdehyde Dehydrogenase From Burkholderia Cenocepacia At 2.38A Resolution
Organism: Burkholderia cenocepacia
Method: ELECTRON MICROSCOPY Release Date: 2025-09-17 Classification: OXIDOREDUCTASE Ligands: NAD |
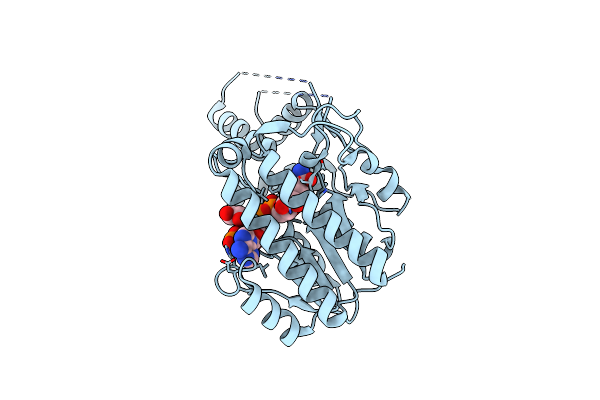 |
Organism: Brucella ovis
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2022-08-10 Classification: ISOMERASE Ligands: NAP, GOL |
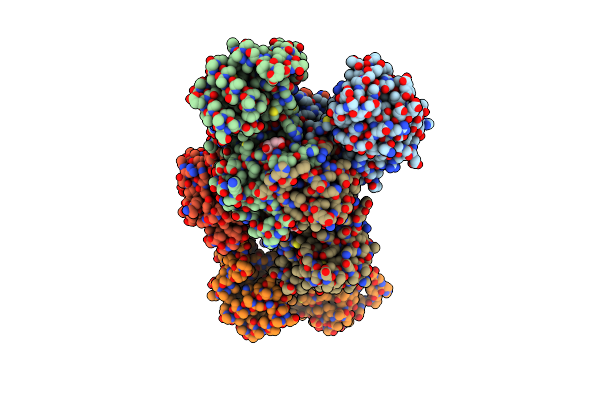 |
Crystal Structure Of Dihydrodipicolinate Reductase From Acinetobacter Baumannii
Organism: Acinetobacter baumannii
Method: X-RAY DIFFRACTION Resolution:2.65 Å Release Date: 2022-06-22 Classification: OXIDOREDUCTASE Ligands: CIT |
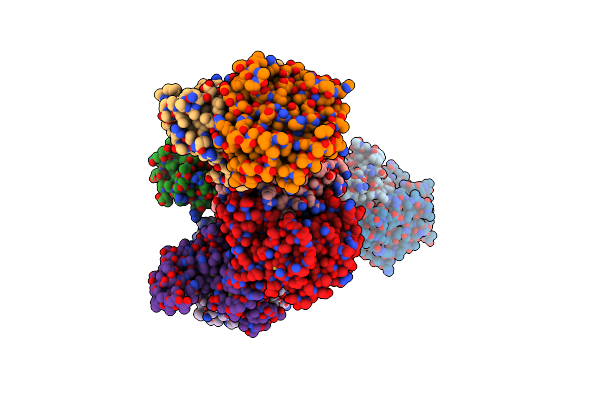 |
Crystal Structure Of Orotidine 5'-Phosphate Decarboxylase From Klebsiella Pneumoniae In Complex With Uridine-5'-Monophosphate
Organism: Klebsiella pneumoniae subsp. pneumoniae hs11286
Method: X-RAY DIFFRACTION Resolution:2.60 Å Release Date: 2022-06-08 Classification: LYASE Ligands: U5P, EDO |
 |
Crystal Structure Of Transcriptional Regulator, Gntr Family, From Brucella Melitensis
Organism: Brucella melitensis atcc 23457
Method: X-RAY DIFFRACTION Resolution:3.00 Å Release Date: 2022-03-09 Classification: DNA BINDING PROTEIN Ligands: ZN, BR |
 |
Crystal Structure Of Ribulose-Phosphate 3-Epimerase From Pseudomonas Aeruginosa
Organism: Pseudomonas aeruginosa (strain atcc 15692 / dsm 22644 / cip 104116 / jcm 14847 / lmg 12228 / 1c / prs 101 / pao1)
Method: X-RAY DIFFRACTION Resolution:2.55 Å Release Date: 2022-03-09 Classification: ISOMERASE Ligands: ZN |
 |
Crystal Structure Of Phosphoserine Aminotransferase From Klebsiella Pneumoniae Subsp. Pneumoniae In Complex With Pyridoxal Phosphate
Organism: Klebsiella pneumoniae subsp. pneumoniae hs11286
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2022-01-12 Classification: TRANSFERASE Ligands: PLP |
 |
Crystal Structure Of Udp-N-Acetylmuramoylalanine--D-Glutamate Ligase (Murd) From Pseudomonas Aeruginosa Pao1
Organism: Pseudomonas aeruginosa (strain atcc 15692 / dsm 22644 / cip 104116 / jcm 14847 / lmg 12228 / 1c / prs 101 / pao1)
Method: X-RAY DIFFRACTION Resolution:2.75 Å Release Date: 2021-12-15 Classification: LIGASE Ligands: ACT, FMT |
 |
Crystal Structure Of A 8-Amino-7-Oxononanoate Synthase/2-Amino-3-Ketobutyrate Coenzyme A Ligase From Mycobacterium Smegmatis
Organism: Mycolicibacterium smegmatis (strain atcc 700084 / mc(2)155)
Method: X-RAY DIFFRACTION Resolution:2.25 Å Release Date: 2021-09-22 Classification: TRANSFERASE |
 |
Translation Initiation Factor Eif-5A Family Protein From Naegleria Fowleri Atcc 30863
Organism: Naegleria fowleri
Method: X-RAY DIFFRACTION Resolution:2.45 Å Release Date: 2021-06-23 Classification: GENE REGULATION |
 |
Crystal Structure Of A Putative Deoxyhypusine Synthase From Entamoeba Histolytica
Organism: Entamoeba histolytica
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2021-02-10 Classification: TRANSFERASE Ligands: CL |
 |
Crystal Structure Of Glutamyl-Trna Synthetase (Gltx) From Stenotrophomonas Maltophilia
Organism: Stenotrophomonas maltophilia (strain k279a)
Method: X-RAY DIFFRACTION Release Date: 2020-10-07 Classification: LIGASE Ligands: ACT, EDO |
 |
Crystal Structure Of Ldha In Complex With Compound Ncgc00384414-01 At 2.05 A Resolution
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.05 Å Release Date: 2020-09-23 Classification: OXIDOREDUCTASE/OXIDOREDUCTASE inhibitor Ligands: PO4, P8M, NAI, GOL |
 |
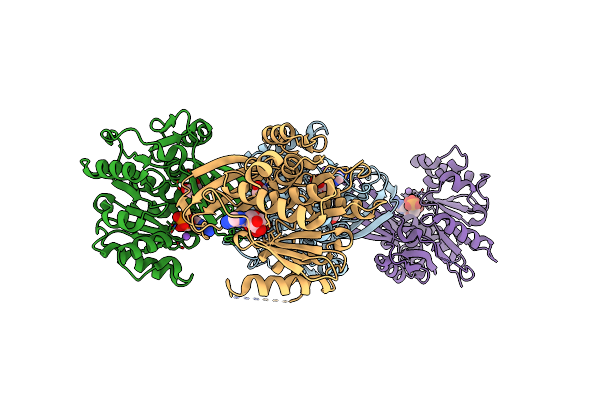
Crystal Structure Of Ldha In Complex With Compound Ncgc00420737-09 At 2.00 A Resolution
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2020-09-23 Classification: OXIDOREDUCTASE/OXIDOREDUCTASE inhibitor Ligands: NAI, P8V, EDO |
 |
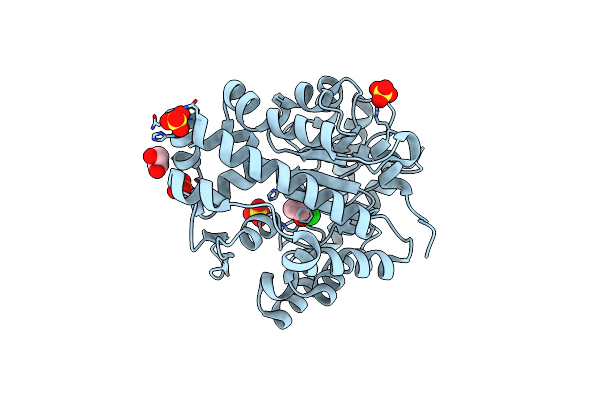
Crystal Structure Of Ribokinase From Cryptococcus Neoformans Var. Grubii Serotype A In Complex With Adp
Organism: Cryptococcus neoformans var. grubii serotype a (strain h99 / atcc 208821 / cbs 10515 / fgsc 9487)
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2020-07-15 Classification: TRANSFERASE Ligands: ADP, EDO, NA |
 |
Crystal Structure Of Uroporphyrinogen Iii Decarboxylate (Heme) From Stenotrophomonas Maltophilia
Organism: Stenotrophomonas maltophilia (strain k279a)
Method: X-RAY DIFFRACTION Resolution:1.55 Å Release Date: 2020-04-22 Classification: LYASE Ligands: EDO, SO4, CL |
 |
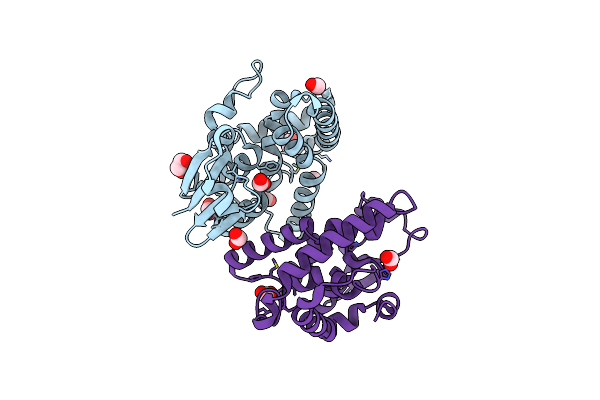
Crystal Structure Of Cystathionine Beta Synthase From Legionella Pneumophila With Llp, Plp, And Homocysteine
Organism: Legionella pneumophila
Method: X-RAY DIFFRACTION Resolution:1.30 Å Release Date: 2020-02-05 Classification: LYASE Ligands: EDO, ACT, HCS, PLP |
 |
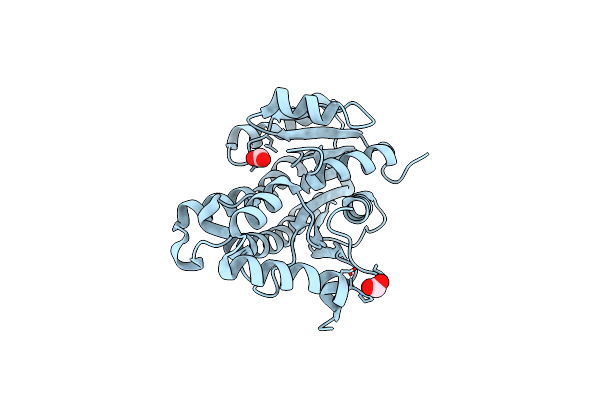
Crystal Structure Of Stringent Starvation Protein A (Bth_I2974) From Burkholderia Thailandensis
Organism: Burkholderia thailandensis e264
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2020-01-15 Classification: STRUCTURAL GENOMICS Ligands: FMT, ACT, EDO |
 |
Crystal Structure Of Enoyl-[Acyl-Carrier-Protein] Reductase [Nadh] (Inha) From Mycobacterium Kansasii
Organism: Mycobacterium kansasii
Method: X-RAY DIFFRACTION Resolution:1.95 Å Release Date: 2019-10-16 Classification: OXIDOREDUCTASE Ligands: FMT |