Search Count: 18
 |
Crystal Structure Of Pi3Kgamma With A Dihydropurinone Inhibitor (Compound 18)
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.74 Å Release Date: 2022-07-13 Classification: SIGNALING PROTEIN Ligands: IGJ |
 |
Human Class I Major Histocompatibility Complex, A02 Allele, Presenting Iigwmwipv
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.01 Å Release Date: 2021-06-30 Classification: IMMUNE SYSTEM Ligands: EDO, GOL, SO4, NA, P4G, PE8, CA |
 |
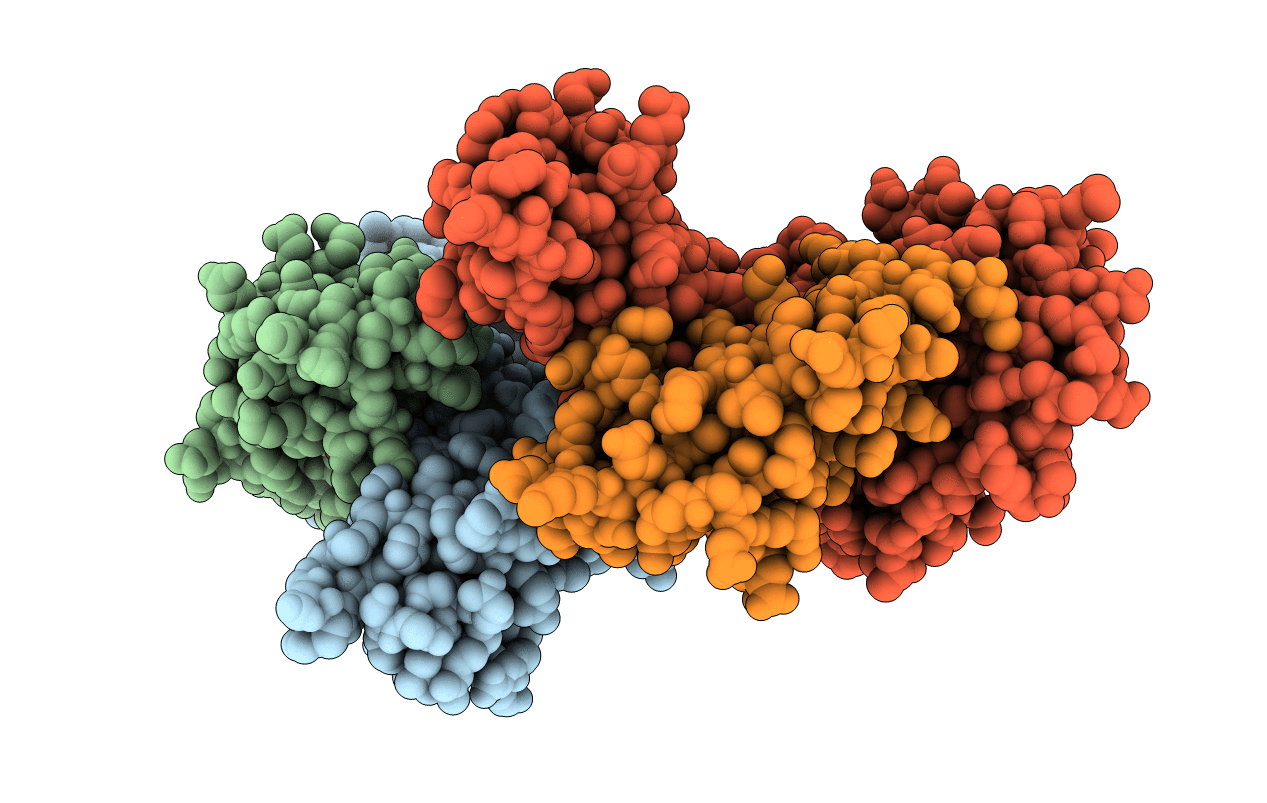
Human Class I Major Histocompatibility Complex, A02 Allele, Presenting Llgwvfaqv
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.70 Å Release Date: 2021-06-30 Classification: IMMUNE SYSTEM |
 |
Human Class I Major Histocompatibility Complex, A02 Allele, Presenting Lls (T-Butyl)Y Fgtpt
Organism: Homo sapiens, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.68 Å Release Date: 2021-06-30 Classification: IMMUNE SYSTEM Ligands: SO4 |
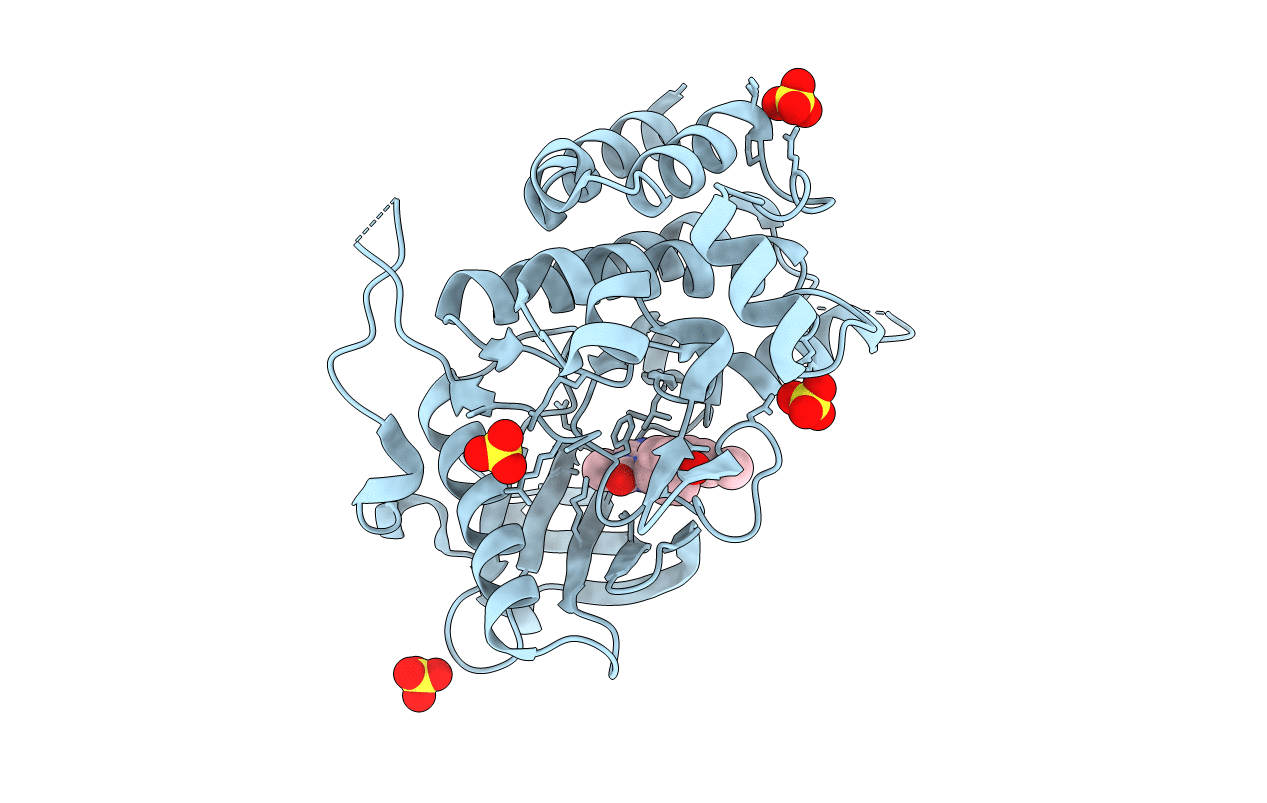 |
Crystal Structure Of The Csf1R Kinase Domain With A Dihydropurinone Inhibitor (Compound 4)
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2020-01-01 Classification: TRANSFERASE Ligands: M9T, SO4 |
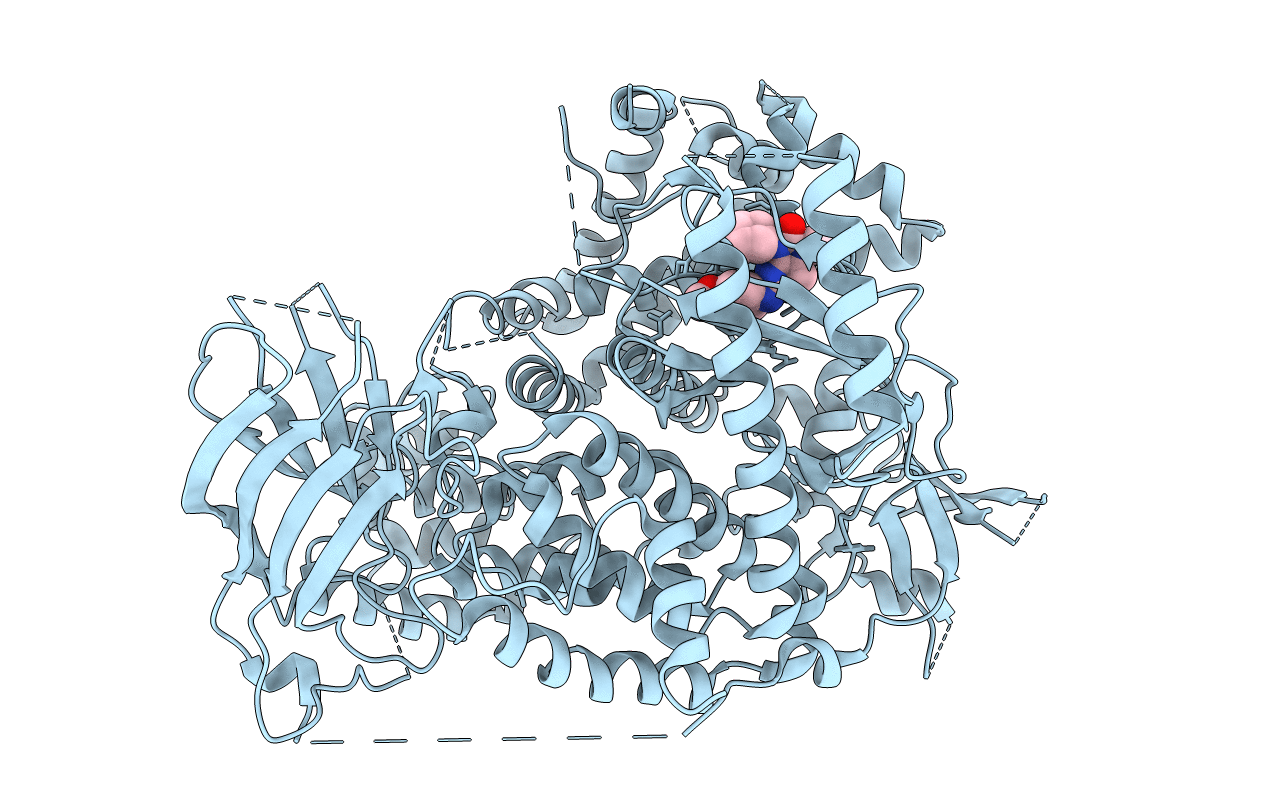 |
Crystal Structure Of Pi3Kgamma With A Dihydropurinone Inhibitor (Compound 4)
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:3.01 Å Release Date: 2020-01-01 Classification: TRANSFERASE Ligands: M9T |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.62 Å Release Date: 2020-01-01 Classification: TRANSFERASE Ligands: MBW |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.49 Å Release Date: 2013-02-27 Classification: TRANSFERASE/TRANSFERASE INHIBITOR Ligands: 0XZ, GOL |
 |
Nmr Structure Of The Histone-Interacting N-Terminal Homodimeric Region Of Rtt106
Organism: Saccharomyces cerevisiae
Method: SOLUTION NMR Release Date: 2012-02-01 Classification: CHAPERONE |
 |
Organism: Saccharomyces cerevisiae
Method: X-RAY DIFFRACTION Resolution:2.59 Å Release Date: 2012-02-01 Classification: CHAPERONE |
 |
Organism: Saccharomyces cerevisiae
Method: X-RAY DIFFRACTION Resolution:1.77 Å Release Date: 2012-02-01 Classification: CHAPERONE Ligands: AHN, GOL |
 |
Organism: Saccharomyces cerevisiae
Method: X-RAY DIFFRACTION Resolution:1.43 Å Release Date: 2009-12-22 Classification: CHAPERONE Ligands: GOL, MLA |
 |
Organism: Saccharomyces cerevisiae
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2009-09-08 Classification: PROTEIN TRANSPORT, ENDOCYTOSIS |
 |
Crystal Structure Of S.Cerevisiae Ist1 N-Terminal Domain In Complex With Did2 Mim Motif
Organism: Saccharomyces cerevisiae
Method: X-RAY DIFFRACTION Resolution:3.80 Å Release Date: 2009-09-08 Classification: PROTEIN TRANSPORT, ENDOCYTOSIS |
 |
Organism: Saccharomyces cerevisiae
Method: X-RAY DIFFRACTION Resolution:2.90 Å Release Date: 2008-01-22 Classification: LIPID TRANSPORT |
 |
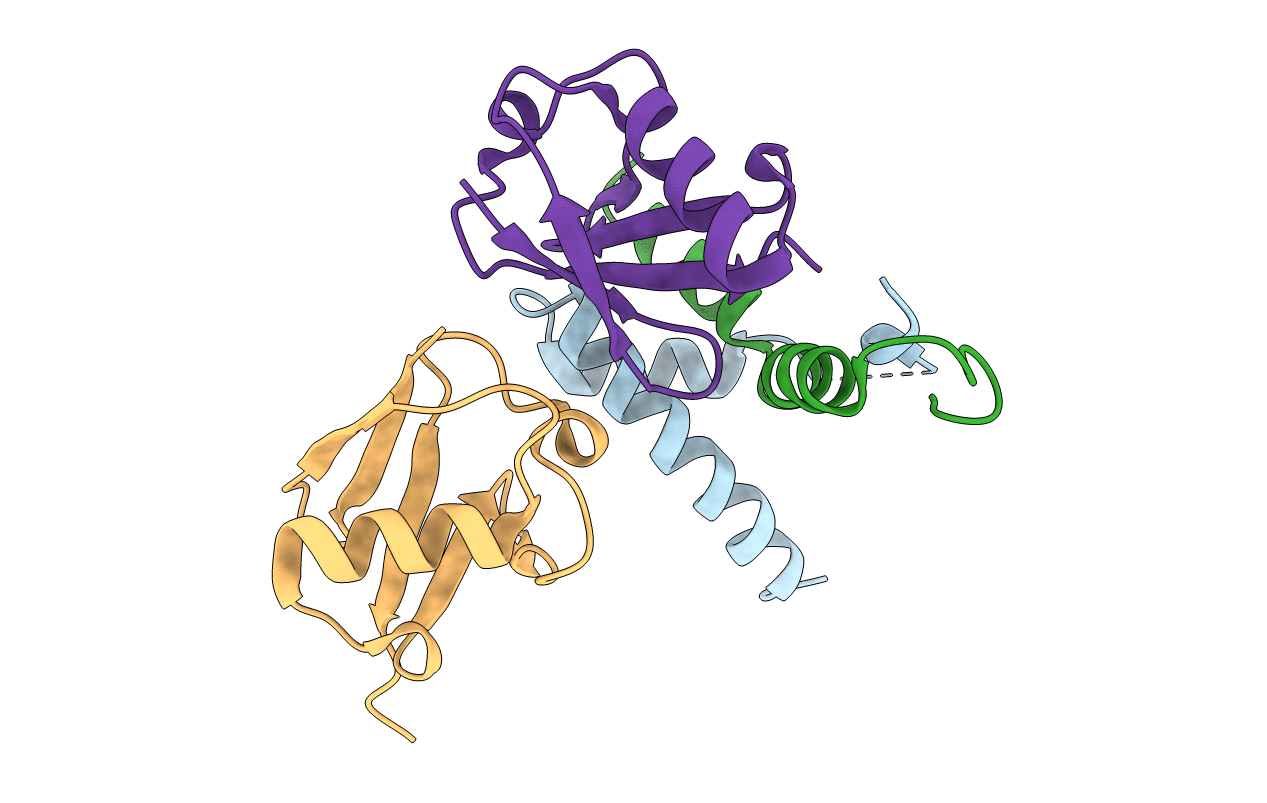
Organism: Saccharomyces cerevisiae
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2008-01-22 Classification: LIPID TRANSPORT Ligands: MPD |
 |
Organism: Saccharomyces cerevisiae, Bos taurus
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2003-06-24 Classification: TRANSLATION |
 |
Organism: Saccharomyces cerevisiae
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2003-06-10 Classification: PROTEIN TRANSPORT |

