Search Count: 44
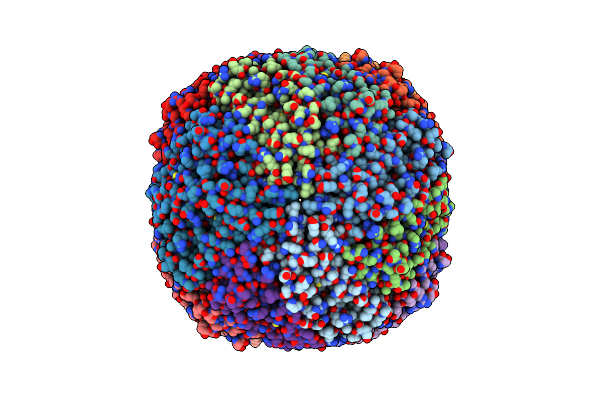 |
Folding Of An Iron Binding Peptide In Response To Sedimentation Is Resolved Using Ferritin As A Nano-Reactor
Organism: Mus musculus
Method: ELECTRON MICROSCOPY Release Date: 2021-07-14 Classification: METAL BINDING PROTEIN Ligands: FE |
 |
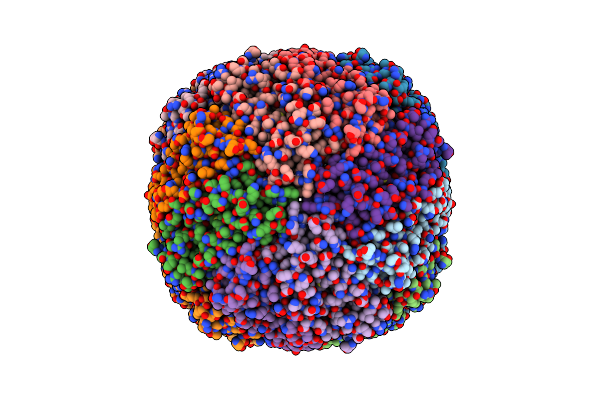
Folding Of An Iron Binding Peptide In Response To Sedimentation Is Resolved Using Ferritin As A Nano-Reactor
Organism: Mus musculus
Method: ELECTRON MICROSCOPY Release Date: 2021-07-07 Classification: METAL BINDING PROTEIN |
 |
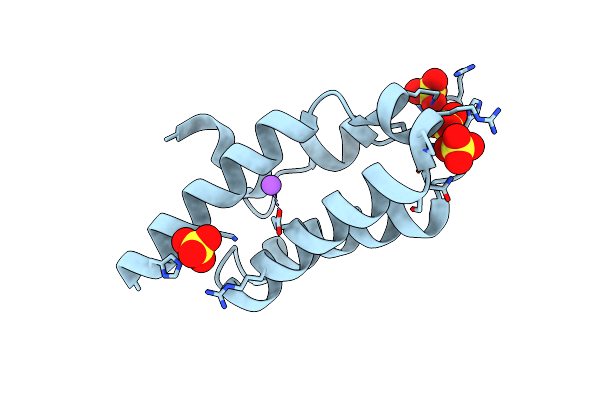
Folding Of An Iron Binding Peptide In Response To Sedimentation Is Resolved Using Ferritin As A Nano-Reactor
Organism: Mus musculus
Method: ELECTRON MICROSCOPY Release Date: 2021-04-28 Classification: METAL BINDING PROTEIN Ligands: FE |
 |
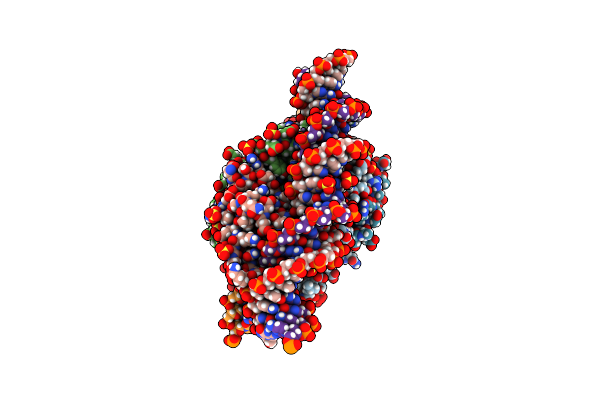
Organism: Mus musculus
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2021-03-31 Classification: METAL BINDING PROTEIN Ligands: ACT |
 |
Organism: Bordetella pertussis
Method: X-RAY DIFFRACTION Resolution:2.01 Å Release Date: 2019-09-18 Classification: LIPID BINDING PROTEIN Ligands: SO4, NA |
 |
Organism: Halobacterium salinarum nrc-1
Method: X-RAY DIFFRACTION Resolution:2.13 Å Release Date: 2019-07-10 Classification: DNA BINDING PROTEIN Ligands: SO4, MN |
 |
Organism: Halobacterium salinarum nrc-1
Method: X-RAY DIFFRACTION Resolution:2.44 Å Release Date: 2019-07-10 Classification: DNA BINDING PROTEIN Ligands: SO4, MN |
 |
Organism: Halobacterium salinarum (strain atcc 700922 / jcm 11081 / nrc-1), Halobacterium salinarum nrc-1
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2019-07-10 Classification: DNA BINDING PROTEIN Ligands: MN, SO4, MES |
 |
Organism: Halobacterium salinarum (strain atcc 700922 / jcm 11081 / nrc-1), Halobacterium salinarum nrc-1
Method: X-RAY DIFFRACTION Resolution:2.68 Å Release Date: 2019-07-10 Classification: DNA BINDING PROTEIN Ligands: MN, SO4 |
 |
Organism: Halobacterium salinarum (strain atcc 700922 / jcm 11081 / nrc-1)
Method: X-RAY DIFFRACTION Resolution:1.75 Å Release Date: 2018-08-22 Classification: DNA BINDING PROTEIN Ligands: BR, SO4 |
 |
Organism: Halobacterium salinarum nrc-1
Method: X-RAY DIFFRACTION Resolution:1.55 Å Release Date: 2018-08-22 Classification: DNA BINDING PROTEIN Ligands: CL, SO4 |
 |
Organism: Halobacterium salinarum (strain atcc 700922 / jcm 11081 / nrc-1)
Method: X-RAY DIFFRACTION Resolution:1.95 Å Release Date: 2018-08-22 Classification: DNA BINDING PROTEIN Ligands: BR, SO4 |
 |
Organism: Halobacterium salinarum (strain atcc 700922 / jcm 11081 / nrc-1)
Method: X-RAY DIFFRACTION Resolution:1.85 Å Release Date: 2018-08-22 Classification: DNA BINDING PROTEIN Ligands: CL, SO4 |
 |
Organism: Escherichia coli o157:h7, Magnetospirillum magneticum amb-1
Method: X-RAY DIFFRACTION Resolution:2.29 Å Release Date: 2017-11-01 Classification: MAGNETITE-BINDING PROTEIN |
 |
Organism: Methanosarcina mazei, Magnetospirillum sp. xm-1
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2017-10-25 Classification: TRANSPORT PROTEIN |
 |
Organism: Magnetospira sp. qh-2
Method: X-RAY DIFFRACTION Resolution:2.53 Å Release Date: 2017-02-01 Classification: SIGNALING PROTEIN Ligands: ZN |
 |
Organism: Magnetospira sp. qh-2
Method: X-RAY DIFFRACTION Resolution:2.15 Å Release Date: 2017-02-01 Classification: METAL TRANSPORT |
 |
Organism: Magnetospira sp. qh-2
Method: X-RAY DIFFRACTION Resolution:1.99 Å Release Date: 2017-02-01 Classification: METAL TRANSPORT Ligands: MG |
 |
Organism: Magnetospira sp. qh-2
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2017-02-01 Classification: METAL TRANSPORT Ligands: MG |
 |
Organism: Magnetospirillum gryphiswaldense
Method: X-RAY DIFFRACTION Resolution:1.79 Å Release Date: 2016-11-30 Classification: METAL TRANSPORT Ligands: SO4 |

