Search Count: 22
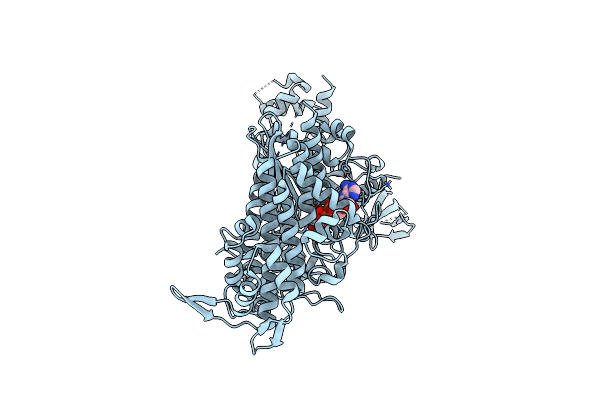 |
Organism: Bos taurus
Method: X-RAY DIFFRACTION Resolution:2.76 Å Release Date: 2023-12-13 Classification: CONTRACTILE PROTEIN Ligands: ADP, VO4, MG |
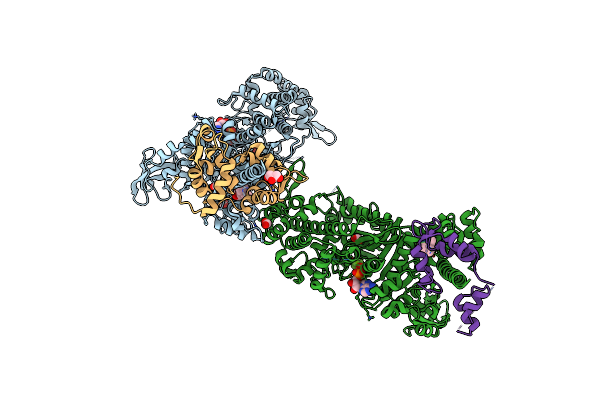 |
Beta-Cardiac Myosin S1 Fragment In The Pre-Powerstroke State Complexed To Mavacamten
Organism: Bos taurus
Method: X-RAY DIFFRACTION Release Date: 2023-12-13 Classification: CONTRACTILE PROTEIN Ligands: ADP, BEF, MG, XB2, FMT, GOL |
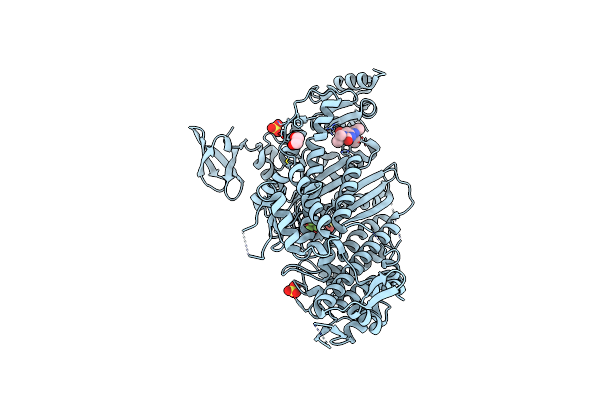 |
Beta-Cardiac Myosin Motor Domain In The Pre-Powerstroke State Complexed To Mavacamten
Organism: Bos taurus
Method: X-RAY DIFFRACTION Release Date: 2023-12-13 Classification: CONTRACTILE PROTEIN Ligands: XB2, SO4, EDO, MG, ADP, BEF |
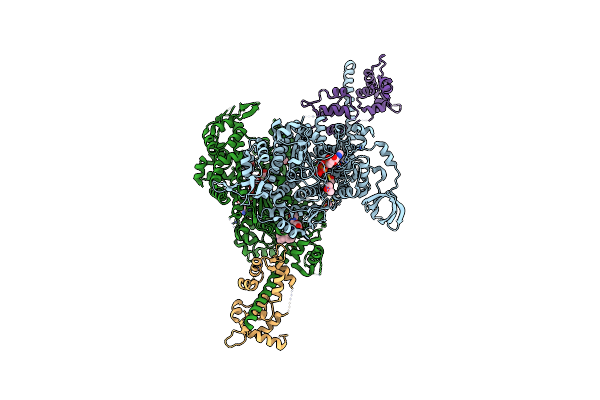 |
Beta-Cardiac Myosin S1 Fragment In The Pre-Powerstroke State Complexed To Omecamtiv Mecarbil
Organism: Bos taurus
Method: X-RAY DIFFRACTION Resolution:1.96 Å Release Date: 2023-12-13 Classification: CONTRACTILE PROTEIN Ligands: ADP, VO4, MG, FMT, DMS, 2OW, GOL, MLA |
 |
Organism: Mus musculus
Method: X-RAY DIFFRACTION Resolution:2.22 Å Release Date: 2023-08-30 Classification: MOTOR PROTEIN |
 |
Organism: Quasibacillus thermotolerans
Method: ELECTRON MICROSCOPY Release Date: 2021-09-29 Classification: VIRUS LIKE PARTICLE |
 |
Crystal Structure Of Cryptosporidium Hominis Cpsf3 In Complex With Compound 61
Organism: Cryptosporidium hominis tu502
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2019-11-20 Classification: RNA BINDING PROTEIN Ligands: ZN, HJB, IPA, GOL, MG |
 |
Organism: Cryptosporidium hominis
Method: X-RAY DIFFRACTION Resolution:1.55 Å Release Date: 2019-11-20 Classification: RNA BINDING PROTEIN Ligands: ZN, GOL, IPA, MG, PG4 |
 |
Extracellulr Xylanase From Geobacillus Stearothermophilus: E159Q Mutant, With Xylopentaose In Active Site
Organism: Geobacillus stearothermophilus
Method: X-RAY DIFFRACTION Resolution:2.01 Å Release Date: 2015-03-18 Classification: HYDROLASE Ligands: ZN, CL |
 |
Extracellulr Xylanase From Geobacillus Stearothermophilus: E159Q Mutant, With Xylotetraose In Active Site
Organism: Geobacillus stearothermophilus
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2015-03-18 Classification: HYDROLASE Ligands: ZN, CL |
 |
Xylanase T6 (Xt6) From Geobacillus Stearothermophilus In Complex With Xylohexaose
Organism: Geobacillus stearothermophilus
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2015-03-11 Classification: HYDROLASE Ligands: ZN, SO4, CL |
 |
High Resolution Crystal Structure Of 1,5-Alpha-L-Arabinanase From Geobacillus Stearothermophilus
Organism: Geobacillus stearothermophilus
Method: X-RAY DIFFRACTION Resolution:1.06 Å Release Date: 2009-04-21 Classification: HYDROLASE Ligands: CA, GOL |
 |
High Resolution Crystal Structure Of 1,5-Alpha-Arabinanase Catalytic Mutant (Abnbe201A)
Organism: Geobacillus stearothermophilus
Method: X-RAY DIFFRACTION Resolution:1.22 Å Release Date: 2009-04-21 Classification: HYDROLASE Ligands: CA |
 |
Crystal Structure Analysis Of 1,5-Alpha-Arabinanase Catalytic Mutant (Abnbe201A) Complexed To Arabinotriose
Organism: Geobacillus stearothermophilus
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2009-04-21 Classification: HYDROLASE Ligands: CA |
 |
Crystal Structure Analysis Of 1,5-Alpha-Arabinanase Catalytic Mutant (D27A)
Organism: Geobacillus stearothermophilus
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2009-04-21 Classification: HYDROLASE Ligands: CA |
 |
Crystal Structure Analysis Of 1,5-Alpha-Arabinanase Catalytic Mutant (Abnbd147A) Complexed To Arabinobiose
Organism: Geobacillus stearothermophilus
Method: X-RAY DIFFRACTION Resolution:1.95 Å Release Date: 2009-04-21 Classification: HYDROLASE Ligands: CA |
 |
Structure Of The Family43 Beta-Xylosidase From Geobacillus Stearothermophilus
Organism: Geobacillus stearothermophilus
Method: X-RAY DIFFRACTION Resolution:1.88 Å Release Date: 2006-04-04 Classification: HYDROLASE Ligands: CA, MES, GOL |
 |
Structure Of The Family43 Beta-Xylosidase D15G Mutant From Geobacillus Stearothermophilus
Organism: Geobacillus stearothermophilus
Method: X-RAY DIFFRACTION Resolution:2.15 Å Release Date: 2006-04-04 Classification: HYDROLASE Ligands: CA, MES, GOL |
 |
Structure Of The Family43 Beta-Xylosidase D128G Mutant From Geobacillus Stearothermophilus In Complex With Xylobiose
Organism: Geobacillus stearothermophilus
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2006-04-04 Classification: HYDROLASE Ligands: CA, MES, GOL |
 |
Structure Of The Family43 Beta-Xylosidase E187G From Geobacillus Stearothermophilus In Complex With Xylobiose
Organism: Geobacillus stearothermophilus
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2006-04-04 Classification: HYDROLASE Ligands: CA, MES, GOL |

