Search Count: 24
 |
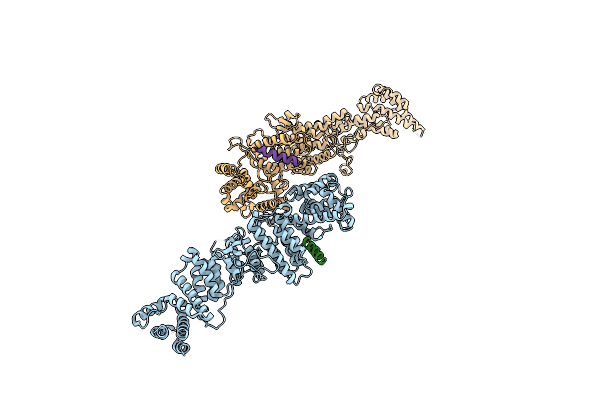
Crystal Structure Of A Peptidergic Gpcr In Complex With A Small Synthetic G Protein-Biased Agonist
Organism: Homo sapiens, Escherichia coli o139:h28 str. e24377a
Method: X-RAY DIFFRACTION Resolution:2.58 Å Release Date: 2024-12-04 Classification: MEMBRANE PROTEIN Ligands: A1D5N, OLA, 1PE |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2017-05-24 Classification: GENE REGULATION Ligands: ZN, GOL, EDO |
 |
Organism: Chaetomium thermophilum (strain dsm 1495 / cbs 144.50 / imi 039719)
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2016-04-20 Classification: TRANSPORT PROTEIN Ligands: PO4, EDO, GOL |
 |
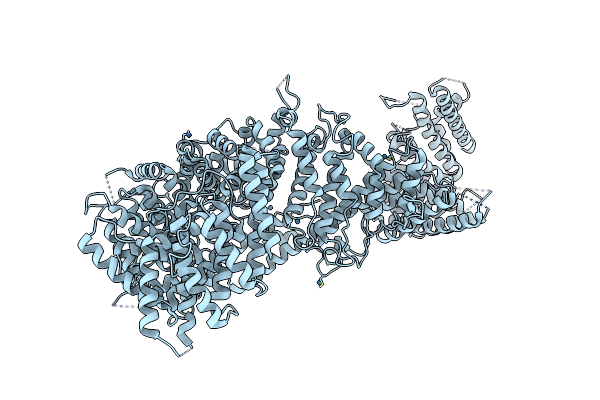
Crystal Structure Of Chaetomium Thermophilum Nup170 Ctd Y905M L1007M L1183M V1292M Mutant
Organism: Chaetomium thermophilum
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2016-04-20 Classification: TRANSPORT PROTEIN |
 |
Organism: Chaetomium thermophilum (strain dsm 1495 / cbs 144.50 / imi 039719)
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2016-04-20 Classification: TRANSPORT PROTEIN Ligands: EDO, EPE, NA |
 |
Organism: Chaetomium thermophilum (strain dsm 1495 / cbs 144.50 / imi 039719)
Method: X-RAY DIFFRACTION Resolution:3.50 Å Release Date: 2016-04-20 Classification: PROTEIN TRANSPORT |
 |
Organism: Chaetomium thermophilum (strain dsm 1495 / cbs 144.50 / imi 039719)
Method: X-RAY DIFFRACTION Resolution:4.01 Å Release Date: 2016-04-20 Classification: TRANSPORT PROTEIN |
 |
Organism: Chaetomium thermophilum (strain dsm 1495 / cbs 144.50 / imi 039719)
Method: X-RAY DIFFRACTION Resolution:3.30 Å Release Date: 2016-04-20 Classification: TRANSPORT PROTEIN |
 |
Organism: Chaetomium thermophilum (strain dsm 1495 / cbs 144.50 / imi 039719)
Method: X-RAY DIFFRACTION Resolution:2.65 Å Release Date: 2016-04-20 Classification: TRANSPORT PROTEIN Ligands: CL |
 |
Organism: Chaetomium thermophilum, Chaetomium thermophilum (strain dsm 1495 / cbs 144.50 / imi 039719)
Method: X-RAY DIFFRACTION Resolution:3.20 Å Release Date: 2016-04-20 Classification: TRANSPORT PROTEIN Ligands: OS |
 |
Organism: Chaetomium thermophilum (strain dsm 1495 / cbs 144.50 / imi 039719)
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2016-04-20 Classification: TRANSPORT PROTEIN Ligands: FLC |
 |
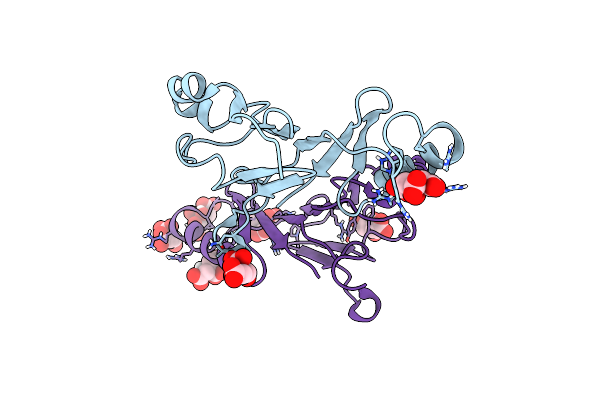
Crystal Structure Of Chaetomium Thermophilum Nup145N Apd T994A Mutant Fused To Nup145C N
Organism: Chaetomium thermophilum (strain dsm 1495 / cbs 144.50 / imi 039719)
Method: X-RAY DIFFRACTION Resolution:1.30 Å Release Date: 2016-04-20 Classification: TRANSPORT PROTEIN Ligands: SO4 |
 |
Organism: Chaetomium thermophilum (strain dsm 1495 / cbs 144.50 / imi 039719)
Method: X-RAY DIFFRACTION Resolution:0.82 Å Release Date: 2016-04-20 Classification: TRANSPORT PROTEIN Ligands: IOD |
 |
Organism: Chaetomium thermophilum (strain dsm 1495 / cbs 144.50 / imi 039719)
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2016-04-20 Classification: TRANSPORT PROTEIN Ligands: SIN, GOL |
 |
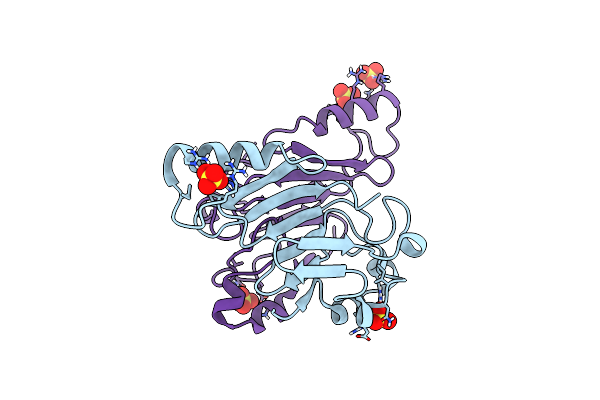
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2014-05-28 Classification: TRANSFERASE Ligands: ACO, CL |
 |
Structure Of Nad-Dependent Protein Deacetylase Sirtuin-1 (Open State, 2.64 A)
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.64 Å Release Date: 2013-11-06 Classification: HYDROLASE Ligands: ZN |
 |
Structure Of Nad-Dependent Protein Deacetylase Sirtuin-1 (Closed State, 2.25 A)
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.25 Å Release Date: 2013-10-23 Classification: HYDROLASE Ligands: ZN, APR |
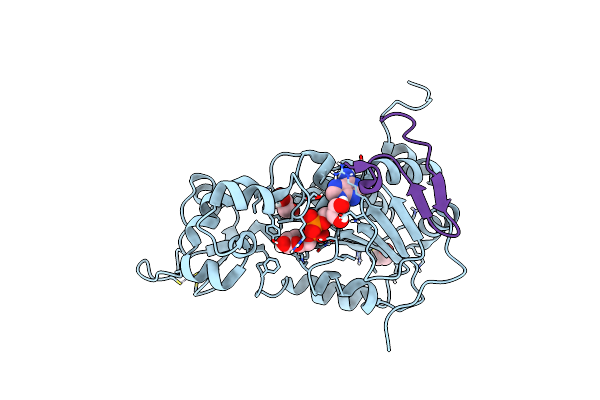 |
Structure Of Nad-Dependent Protein Deacetylase Sirtuin-1 (Closed State, 1.85 A)
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.85 Å Release Date: 2013-10-23 Classification: HYDROLASE Ligands: ZN, APR, BME, GOL |
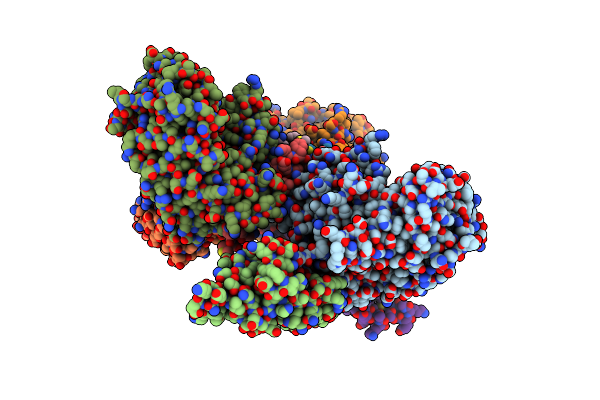 |
Organism: Saccharomyces cerevisiae, Mus musculus
Method: X-RAY DIFFRACTION Resolution:3.40 Å Release Date: 2012-04-11 Classification: PROTEIN TRANSPORT |
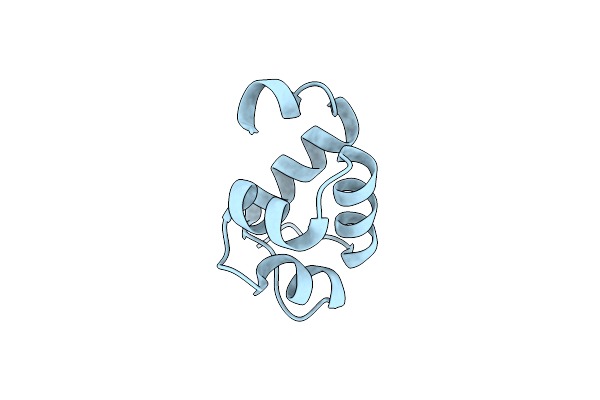 |
Structure Of The C-Terminal Domain Of The E. Coli Protein Mqsa (Ygit/B3021)
Organism: Escherichia coli k-12
Method: X-RAY DIFFRACTION Resolution:1.40 Å Release Date: 2010-01-12 Classification: DNA BINDING PROTEIN |

