Search Count: 24
 |
Cryo-Em Structure Of The Ct-Pt-Bccp Domain Of Pyruvate Carboxylase From Mycobacterium Tuberculosis
Organism: Mycobacterium tuberculosis h37rv
Method: ELECTRON MICROSCOPY Release Date: 2025-05-14 Classification: LIGASE Ligands: ZN, BTN |
 |
Cryo-Em Structure Of Pyruvate Carboxylase From Mycobacterium Tuberculosis In Complex With Acetyl-Coa And Adp
Organism: Mycobacterium tuberculosis h37rv
Method: ELECTRON MICROSCOPY Release Date: 2025-05-07 Classification: LIGASE Ligands: ADP, ZN, ACO |
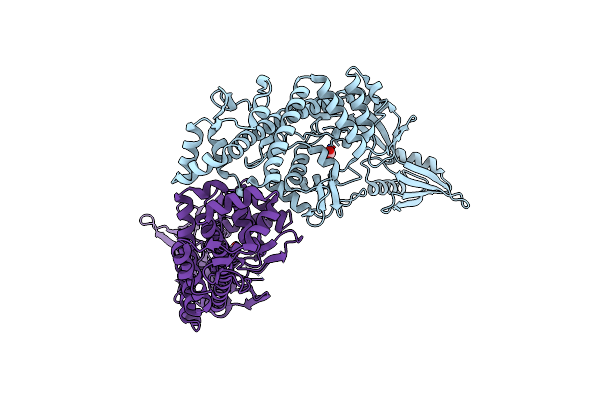 |
Organism: Salmonella enterica subsp. enterica serovar typhi str. ct18
Method: X-RAY DIFFRACTION Release Date: 2024-05-08 Classification: ISOMERASE Ligands: CO3 |
 |
Organism: Mycobacterium tuberculosis h37rv
Method: X-RAY DIFFRACTION Resolution:2.24 Å Release Date: 2022-05-18 Classification: SUGAR BINDING PROTEIN Ligands: SO4 |
 |
Crystal Structure Lpqy In Complex With Trehalose From Mycobacterium Tuberculosis
Organism: Mycobacterium tuberculosis h37rv
Method: X-RAY DIFFRACTION Resolution:1.91 Å Release Date: 2022-05-18 Classification: SUGAR BINDING PROTEIN Ligands: SO4 |
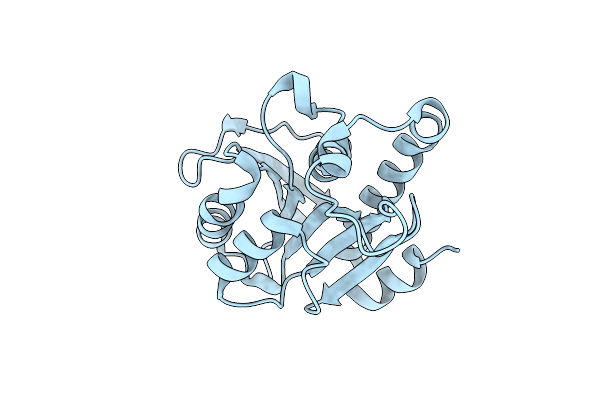 |
Crystal Structure Of Full-Length Peptidyl-Trna Hydrolase From Mycobacterium Tuberculosis
Organism: Mycobacteriaceae bacterium
Method: X-RAY DIFFRACTION Resolution:1.94 Å Release Date: 2022-04-13 Classification: HYDROLASE |
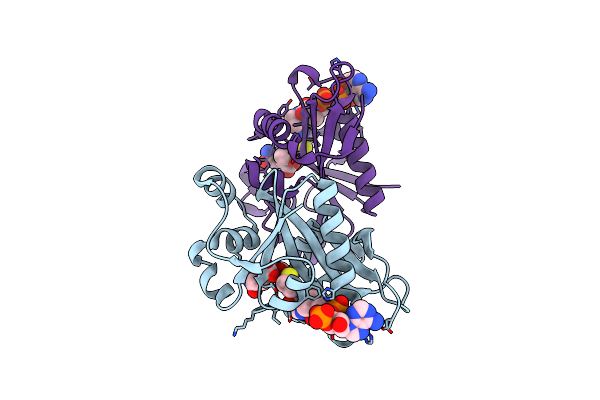 |
The Crystal Structure Of Rv2747 From Mycobacterium Tuberculosis In Complex With Coa And Nlq
Organism: Mycobacterium tuberculosis h37rv
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2018-12-26 Classification: TRANSFERASE Ligands: COA, NLQ |
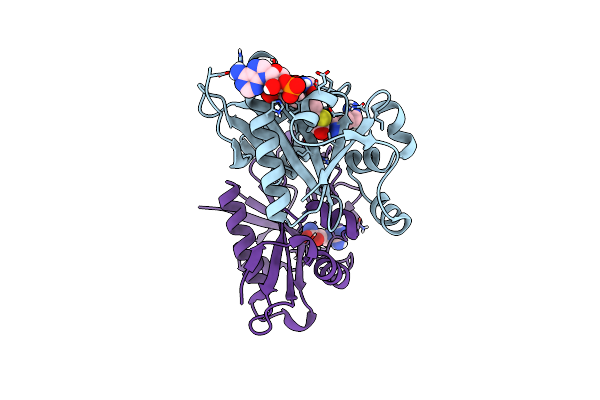 |
The Crystal Structure Of Rv2747 From Mycobacterium Tuberculosis In Complex With Acetyl Coa And L-Arginine
Organism: Mycobacterium tuberculosis h37rv
Method: X-RAY DIFFRACTION Resolution:3.00 Å Release Date: 2018-11-07 Classification: TRANSFERASE Ligands: ARG, ACO |
 |
Organism: Mycobacterium tuberculosis
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2014-11-12 Classification: TOXIN/ANTITOXIN Ligands: MN, MG |
 |
Organism: Ruminiclostridium thermocellum
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2014-10-08 Classification: TRANSFERASE/RNA Ligands: GTP, MG, PO4, NA |
 |
Organism: Ruminiclostridium thermocellum
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2014-10-08 Classification: TRANSFERASE/DNA Ligands: GTP, MG |
 |
Structure Of Bacterial Polynucleotide Kinase Product Complex Bound To Gdp And Dna
Organism: Clostridium thermocellum
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2013-11-06 Classification: TRANSFERASE/DNA Ligands: GDP, MG |
 |
Structure Of Bacterial Polynucleotide Kinase Michaelis Complex Bound To Gtp And Dna
Organism: Clostridium thermocellum
Method: X-RAY DIFFRACTION Resolution:1.73 Å Release Date: 2013-11-06 Classification: TRANSFERASE/DNA Ligands: GTP, MG, CIT |
 |
Organism: Clostridium thermocellum
Method: X-RAY DIFFRACTION Resolution:2.03 Å Release Date: 2013-08-28 Classification: TRANSFERASE Ligands: MG, UTP, NA |
 |
Organism: Clostridium thermocellum
Method: X-RAY DIFFRACTION Resolution:2.14 Å Release Date: 2013-08-28 Classification: TRANSFERASE Ligands: GTP, MG |
 |
Organism: Clostridium thermocellum
Method: X-RAY DIFFRACTION Resolution:2.49 Å Release Date: 2013-08-28 Classification: TRANSFERASE Ligands: MG, CTP |
 |
Organism: Clostridium thermocellum
Method: X-RAY DIFFRACTION Resolution:2.01 Å Release Date: 2013-08-28 Classification: TRANSFERASE Ligands: MG, DTP, NA |
 |
Organism: Clostridium thermocellum
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2012-11-14 Classification: TRANSFERASE Ligands: ADP, MG |
 |
Organism: Clostridium thermocellum
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2012-11-14 Classification: TRANSFERASE Ligands: ATP, MG, CIT, NA |
 |
Cobalt Bound Structure Of An Archaeal Member Of The Ligd 3'-Phosphoesterase Dna Repair Enzyme Family
Organism: Candidatus korarchaeum cryptofilum
Method: X-RAY DIFFRACTION Resolution:1.52 Å Release Date: 2011-10-12 Classification: HYDROLASE Ligands: CO, PO4, PEG |

