Search Count: 193
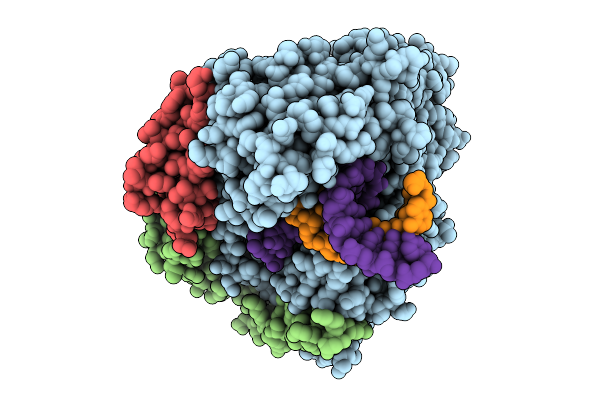 |
Cryo-Em Structure Of Sars Cov-2 Rdrp S759A Mutant In Complex With 20-40Mer Rna Incorporating Remdesivir
Organism: Severe acute respiratory syndrome coronavirus 2
Method: ELECTRON MICROSCOPY Release Date: 2025-10-01 Classification: VIRAL PROTEIN Ligands: ZN |
 |
Cryo-Em Structure Of Sars Cov-2 Rdrp Wild-Type In Complex With 20-40Mer Rna Incorporating Remdesivir
Organism: Severe acute respiratory syndrome coronavirus 2
Method: ELECTRON MICROSCOPY Release Date: 2025-10-01 Classification: VIRAL PROTEIN Ligands: ZN |
 |
Cryo-Em Structure Of Sars Cov-2 Rdrp S759A Mutant In Complex With 20-40Mer Rna Incorporating Atp
Organism: Severe acute respiratory syndrome coronavirus 2
Method: ELECTRON MICROSCOPY Release Date: 2025-10-01 Classification: VIRAL PROTEIN Ligands: ZN |
 |
Cryo-Em Structure Of Sars Cov-2 Rdrp Wild-Type In Complex With 20-40Mer Rna Incorporating Atp
Organism: Severe acute respiratory syndrome coronavirus 2
Method: ELECTRON MICROSCOPY Release Date: 2025-10-01 Classification: VIRAL PROTEIN Ligands: ZN |
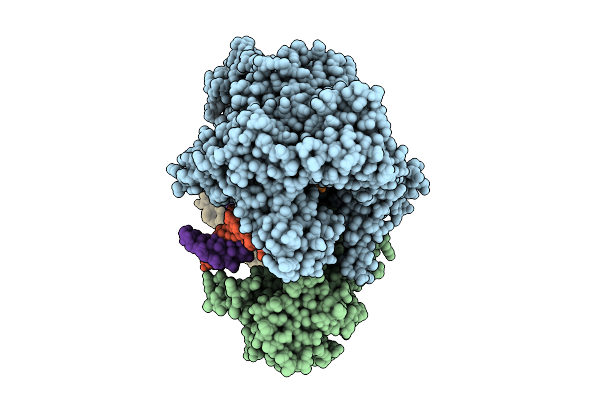 |
Cryo-Em Structure Of The Human Mitochondrial Rna Polymerase Transcription Initiation Complex (Polrmt/Tfam/Tfb2M/Dna/Rna) With A 2-Mer Rna (Pppgpa) And Gtp Poised For Catalysis (Pre-Ic3)
Organism: Homo sapiens, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2025-07-30 Classification: TRANSCRIPTION Ligands: GTP, MG |
 |
Cryo-Em Structure Of The Human Mitochondrial Rna Polymerase Transcription Initiation Complex (Polrmt/Tfb2M/Dna/Rna) Without Tfam; And With A 2-Mer Rna (Pppgpa) And Gtp Poised For Catalysis (Pre-Ic3-Tfam)
Organism: Homo sapiens, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2025-07-30 Classification: TRANSCRIPTION Ligands: GTP, MG |
 |
Cryo-Em Structure Of The Human Mitochondrial Rna Polymerase Transcription Initiation Complex (Polrmt/Tfb2M/Dna/Rna) Without Tfam; And With A Slipped 3-Mer Rna, Pppgpgpa (Slipped Ic3-Tfam)
Organism: Homo sapiens, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2025-07-30 Classification: TRANSCRIPTION |
 |
Cryo-Em Structure Of The Human Mitochondrial Rna Polymerase Transcription Initiation Complex (Polrmt/Tfam/Tfb2M/Dna/Rna) With A Slipped 3-Mer Rna, Pppgpgpa (Slipped Ic3)
Organism: Homo sapiens, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2025-07-30 Classification: TRANSCRIPTION |
 |
Cryo-Em Structure Of The Human Mitochondrial Rna Polymerase Transcription Initiation Complex (Polrmt/Tfam/Tfb2M/Dna/Rna) With A Slipped 3-Mer Rna (Pppgpgpa) And Gtp Poised For Catalysis (Slipped Pre-Ic4)
Organism: Homo sapiens, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2025-07-30 Classification: TRANSCRIPTION Ligands: GTP, MG |
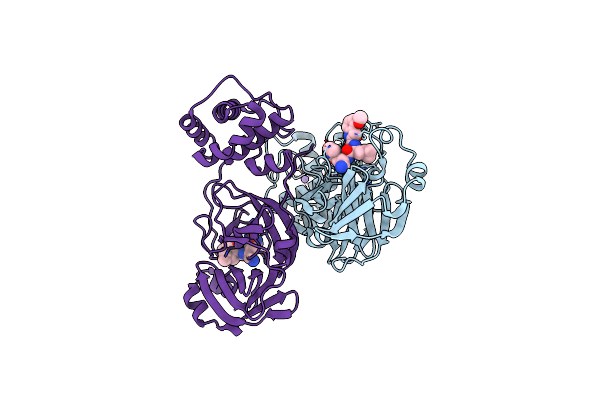 |
Organism: Severe acute respiratory syndrome coronavirus 2
Method: X-RAY DIFFRACTION Release Date: 2025-04-30 Classification: VIRAL PROTEIN, HYDROLASE/INHIBITOR Ligands: A1BCZ, NA |
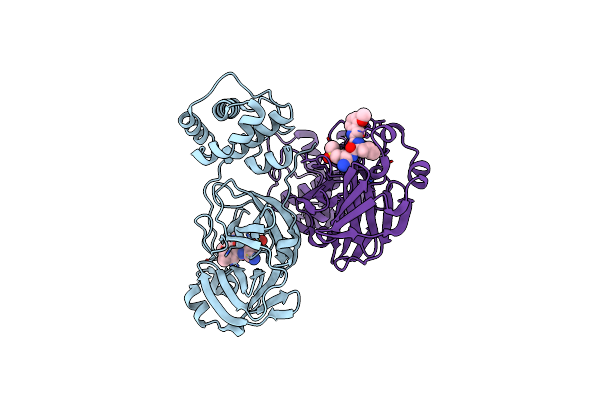 |
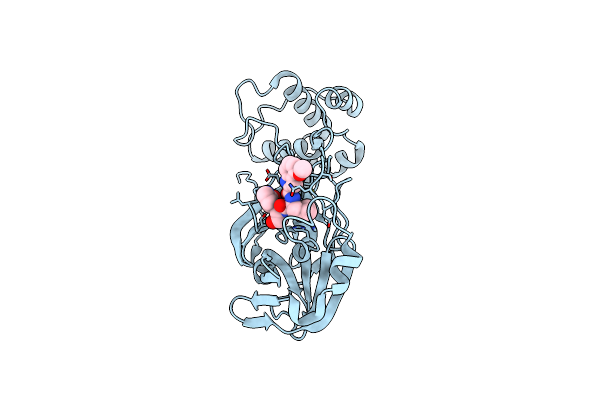
Organism: Severe acute respiratory syndrome coronavirus 2
Method: X-RAY DIFFRACTION Release Date: 2025-04-30 Classification: VIRAL PROTEIN, HYDROLASE/INHIBITOR Ligands: A1BCY |
 |
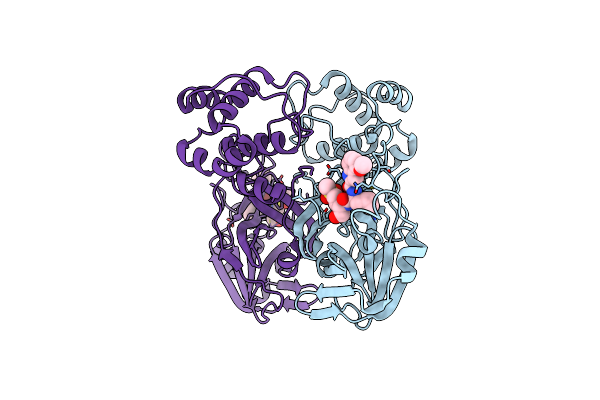
Organism: Severe acute respiratory syndrome coronavirus 2
Method: X-RAY DIFFRACTION Release Date: 2025-04-30 Classification: VIRAL PROTEIN, HYDROLASE/INHIBITOR Ligands: A1BCX |
 |
Organism: Severe acute respiratory syndrome coronavirus 2
Method: X-RAY DIFFRACTION Release Date: 2025-04-30 Classification: VIRAL PROTEIN, HYDROLASE/INHIBITOR Ligands: A1BCW |
 |
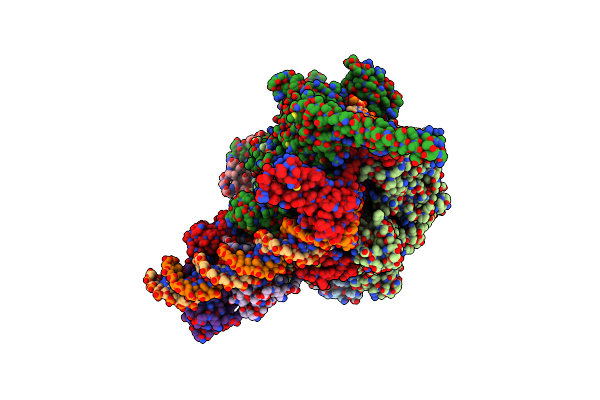
Cryo-Em Structure Of Mycobacterium Tuberculosis Transcription Activation Complex With Unphosphated Phop
Organism: Mycobacterium tuberculosis
Method: ELECTRON MICROSCOPY Release Date: 2025-03-05 Classification: GENE REGULATION Ligands: ZN, MG |
 |
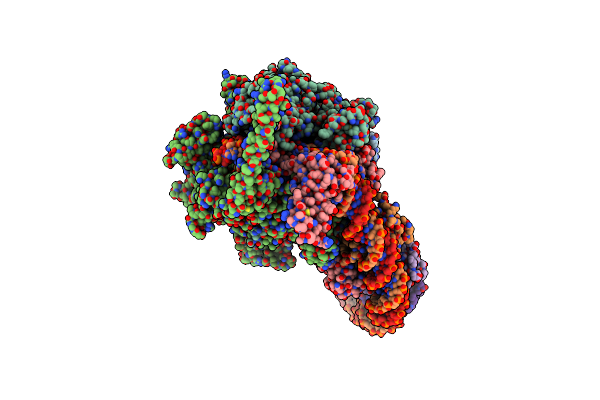
Cryo-Em Structure Of Mycobacterium Tuberculosis Transcription Activation Complex With Two Phop Molecules(Composite Map)
Organism: Mycobacterium tuberculosis h37rv
Method: ELECTRON MICROSCOPY Release Date: 2025-03-05 Classification: GENE REGULATION/DNA Ligands: ZN, MG |
 |
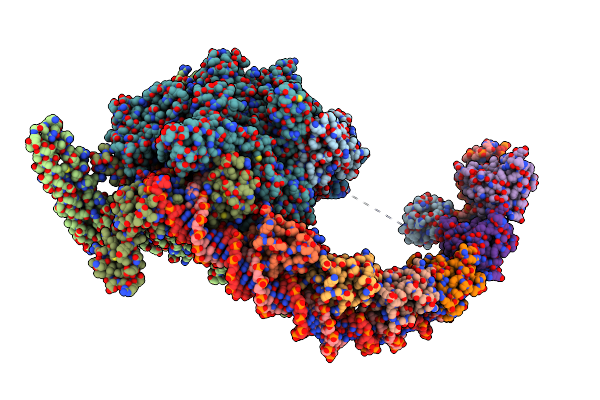
Cryo-Em Structure Of Mycobacterium Tuberculosis Transcription Activation Complex With Four Phop Molecules (Composite Map)
Organism: Mycobacterium tuberculosis h37rv
Method: ELECTRON MICROSCOPY Release Date: 2025-03-05 Classification: GENE REGULATION/DNA Ligands: ZN, MG |
 |
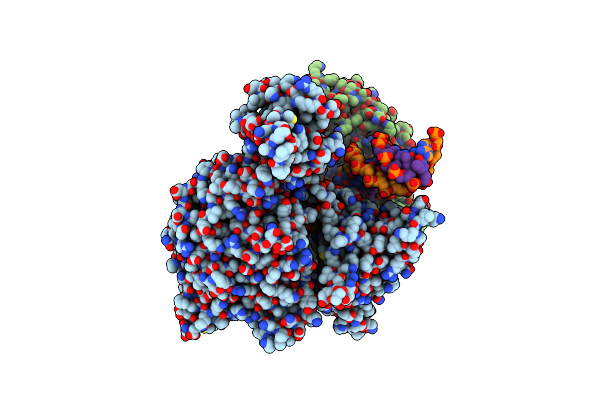
Cryo-Em Structure Of Mycobacterium Tuberculosis Transcription Activation Complex With Six Phop Molecules (Composite Map)
Organism: Mycobacterium tuberculosis h37rv
Method: ELECTRON MICROSCOPY Release Date: 2025-03-05 Classification: GENE REGULATION/DNA Ligands: ZN, MG |
 |
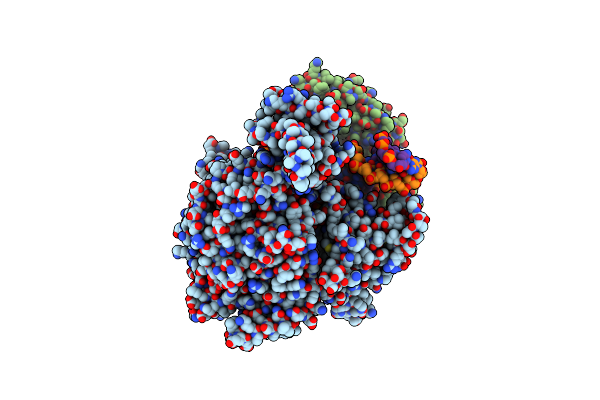
Cryo-Em Structure Of Yeast Mitochondrial Rna Polymerase Transcription Initiation Complex With 4-Mer Rna, Pppgpgpupa (Ic4)
Organism: Saccharomyces cerevisiae s288c
Method: ELECTRON MICROSCOPY Release Date: 2023-08-30 Classification: TRANSCRIPTION Ligands: GTP |
 |
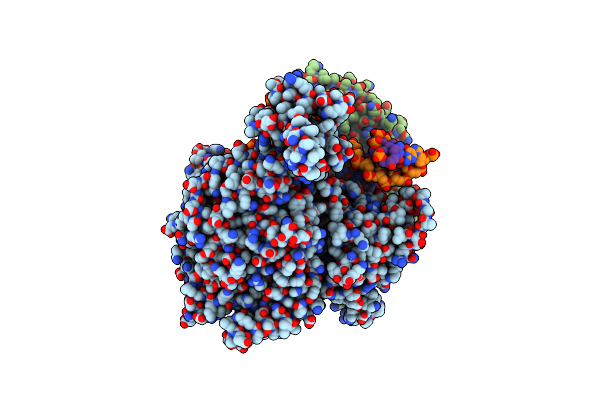
Cryo-Em Structure Of Yeast Mitochondrial Rna Polymerase Transcription Initiation Complex With 5-Mer Rna, Pppgpgpapapa (Ic5)
Organism: Saccharomyces cerevisiae s288c
Method: ELECTRON MICROSCOPY Release Date: 2023-08-30 Classification: TRANSCRIPTION Ligands: GTP |
 |
Cryo-Em Structure Of Yeast Mitochondrial Rna Polymerase Transcription Initiation Complex With 6-Mer Rna, Pppgpgpapapapu (Ic6)
Organism: Saccharomyces cerevisiae s288c
Method: ELECTRON MICROSCOPY Release Date: 2023-08-30 Classification: TRANSCRIPTION Ligands: GTP |

