Search Count: 14
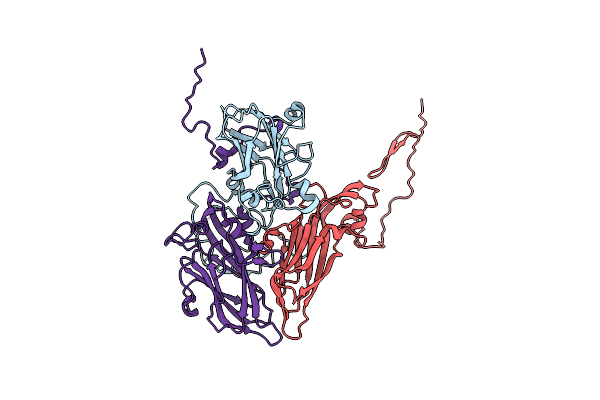 |
Organism: Hepatovirus a, Hepatitis a virus
Method: ELECTRON MICROSCOPY Release Date: 2017-01-25 Classification: VIRUS |
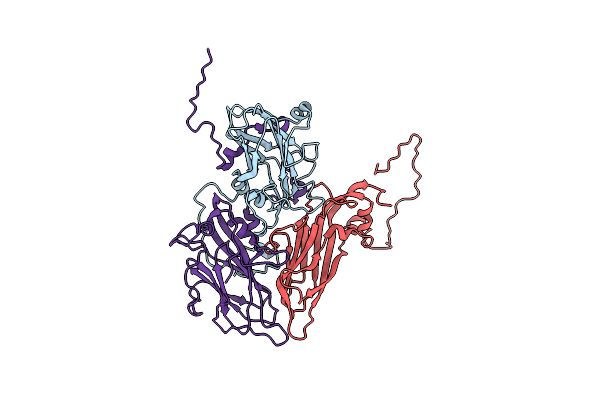 |
Organism: Hepatitis a virus
Method: ELECTRON MICROSCOPY Release Date: 2017-01-25 Classification: VIRUS |
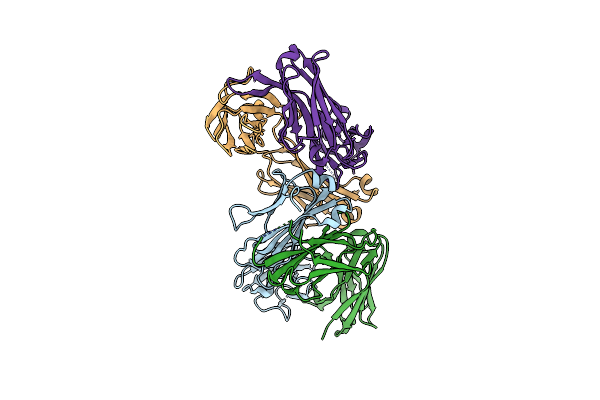 |
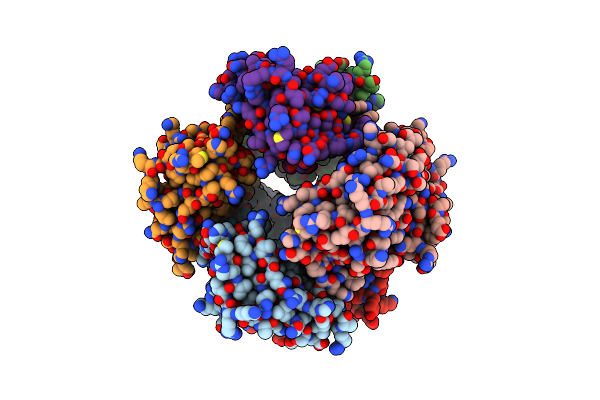
Organism: Mus musculus
Method: X-RAY DIFFRACTION Resolution:2.91 Å Release Date: 2017-01-25 Classification: IMMUNE SYSTEM |
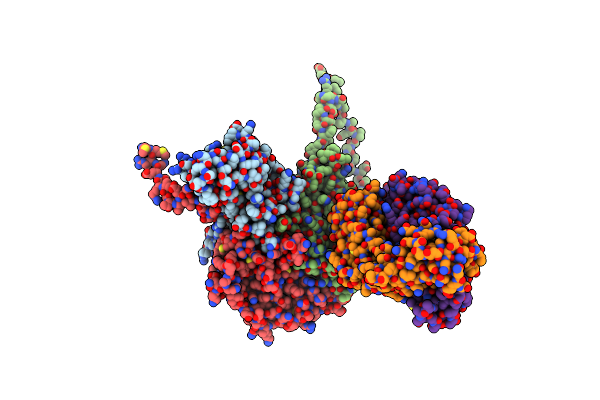 |
Organism: Hepatovirus a, Hepatitis a virus, Mus musculus
Method: ELECTRON MICROSCOPY Release Date: 2017-01-25 Classification: VIRUS |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2015-11-25 Classification: IMMUNE SYSTEM |
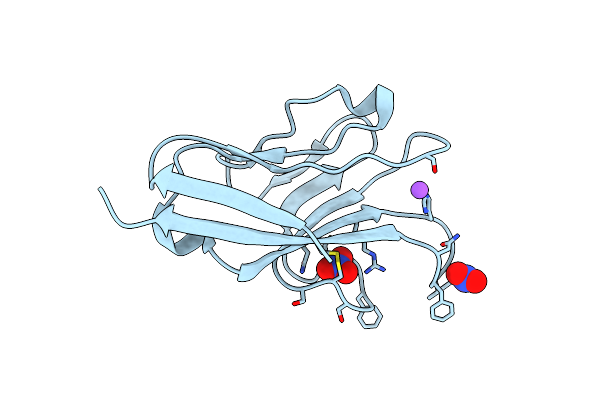 |
Crystal Structure Of Human T-Cell Immunoglobulin And Mucin Domain Protein 1
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.30 Å Release Date: 2015-11-25 Classification: IMMUNE SYSTEM Ligands: NO3, NA |
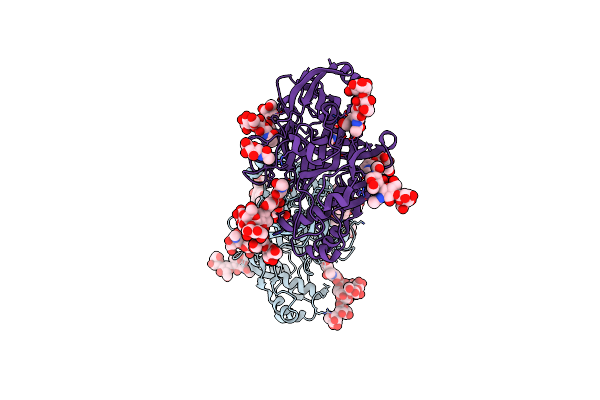 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:3.01 Å Release Date: 2015-07-08 Classification: PROTEIN BINDING Ligands: NAG |
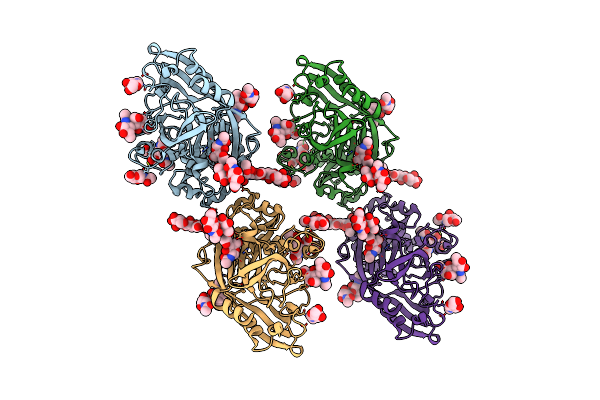 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:3.65 Å Release Date: 2015-07-08 Classification: PROTEIN BINDING Ligands: NAG |
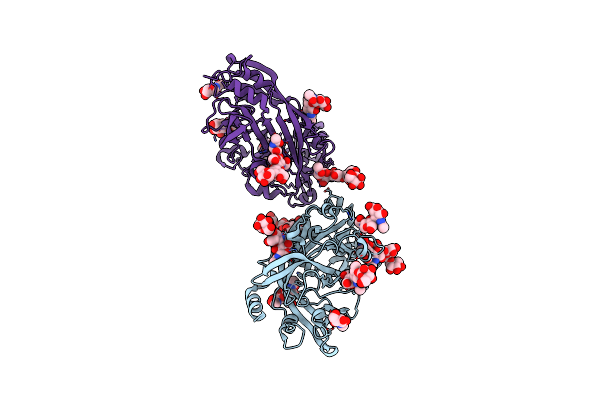 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.89 Å Release Date: 2015-07-08 Classification: PROTEIN BINDING Ligands: NAG |
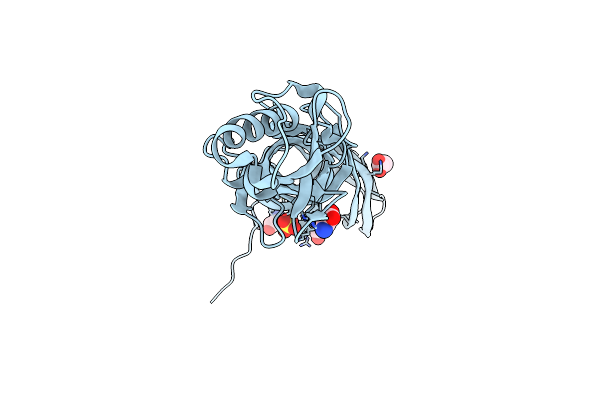 |
Crystal Structure Of Bacillus Thuringiensis Cry51Aa1 Protoxin At 1.65 Angstroms Resolution
Organism: Bacillus thuringiensis
Method: X-RAY DIFFRACTION Resolution:1.65 Å Release Date: 2015-06-03 Classification: TOXIN Ligands: GOL, SO4, GLY |
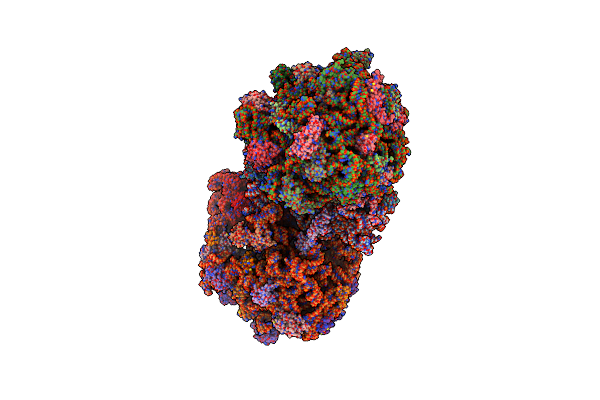 |
Organism: Thermus thermophilus, Escherichia coli
Method: X-RAY DIFFRACTION Resolution:3.90 Å Release Date: 2014-07-09 Classification: RIBOSOME Ligands: MG, PAR, ZN |
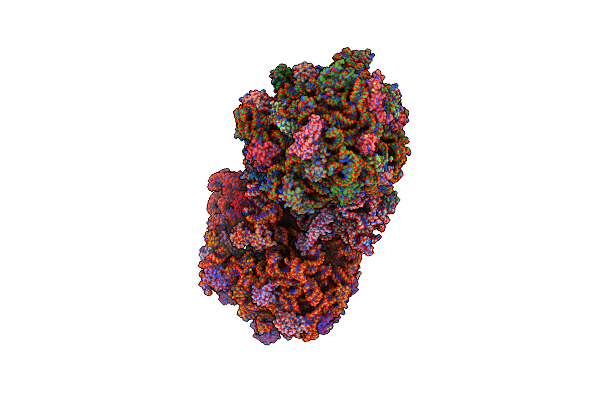 |
Organism: Thermus thermophilus, Escherichia coli
Method: X-RAY DIFFRACTION Resolution:3.52 Å Release Date: 2014-07-09 Classification: RIBOSOME Ligands: MG, PAR, ZN |
 |
Organism: Coxsackievirus a16
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2014-03-26 Classification: HYDROLASE Ligands: ZN, 1PE, TRS |
 |
Organism: Pseudomonas putida
Method: SOLUTION NMR Release Date: 2011-02-16 Classification: OXIDOREDUCTASE Ligands: CAM, HEM, CMO, K, CL |

