Search Count: 62
 |
Organism: Caenorhabditis elegans
Method: X-RAY DIFFRACTION Release Date: 2024-11-27 Classification: RNA BINDING PROTEIN |
 |
Escherichia Coli Rna Polymerase Unwinding Intermediate (I1A) At The Lambda Pr Promoter
Organism: Escherichia coli (strain k12), Escherichia phage lambda
Method: ELECTRON MICROSCOPY Release Date: 2024-07-03 Classification: transcription/DNA Ligands: 4QM, MG, ZN |
 |
Escherichia Coli Rna Polymerase Unwinding Intermediate (I1D) At The Lambda Pr Promoter
Organism: Escherichia coli (strain k12), Escherichia phage lambda
Method: ELECTRON MICROSCOPY Release Date: 2024-07-03 Classification: transcription/DNA Ligands: 4QM, MG, ZN |
 |
Escherichia Coli Rna Polymerase Unwinding Intermediate (I1B) At The Lambda Pr Promoter
Organism: Escherichia coli (strain k12), Escherichia coli k-12, Escherichia phage lambda
Method: ELECTRON MICROSCOPY Release Date: 2024-07-03 Classification: transcription/DNA Ligands: 4QM, MG, ZN |
 |
Escherichia Coli Rna Polymerase Unwinding Intermediate (I1C) At The Lambda Pr Promoter
Organism: Escherichia coli (strain k12), Escherichia phage lambda
Method: ELECTRON MICROSCOPY Release Date: 2024-07-03 Classification: transcription/DNA Ligands: 1N7, MG, ZN |
 |
Escherichia Coli Rna Polymerase Closed Complex Intermediate At The Lambda Pr Promoter
Organism: Escherichia coli (strain k12), Escherichia phage lambda
Method: ELECTRON MICROSCOPY Release Date: 2024-07-03 Classification: transcription/DNA Ligands: 1N7, MG, ZN |
 |
Organism: Thermochaetoides thermophila dsm 1495
Method: ELECTRON MICROSCOPY Release Date: 2023-10-25 Classification: TRANSCRIPTION Ligands: ADP |
 |
Organism: Thermochaetoides thermophila dsm 1495
Method: ELECTRON MICROSCOPY Release Date: 2023-10-25 Classification: TRANSCRIPTION |
 |
Organism: Chaetomium thermophilum, Escherichia coli
Method: X-RAY DIFFRACTION Resolution:3.01 Å Release Date: 2023-10-25 Classification: TRANSCRIPTION/RNA Ligands: ADP, SO4 |
 |
Organism: Severe acute respiratory syndrome coronavirus 2, Camelus dromedarius
Method: ELECTRON MICROSCOPY Release Date: 2022-04-20 Classification: VIRAL PROTEIN/IMMUNE SYSTEM |
 |
Organism: Chaetomium thermophilum, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2022-03-02 Classification: RIBOSOMAL PROTEIN Ligands: ATP, MG, ADP |
 |
Organism: Chaetomium thermophilum, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2022-03-02 Classification: RIBOSOMAL PROTEIN Ligands: ATP, MG, ADP |
 |
Organism: Chaetomium thermophilum, Escherichia coli
Method: ELECTRON MICROSCOPY Release Date: 2022-03-02 Classification: RIBOSOMAL PROTEIN Ligands: ATP, PO4 |
 |
Structure Of Positive Allosteric Modulator-Bound Active Human Calcium-Sensing Receptor
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2022-01-19 Classification: MEMBRANE PROTEIN Ligands: NAG, TCR, PO4, CA, 9IG, CLR |
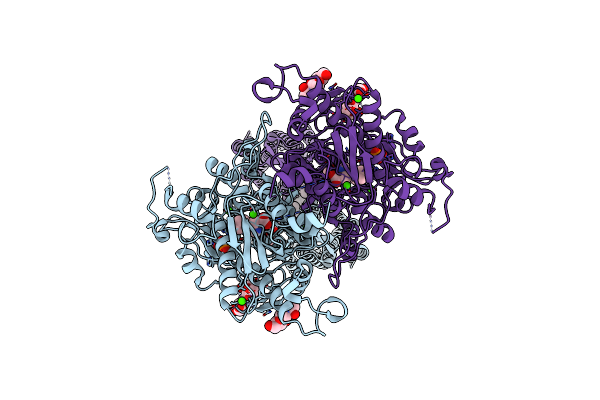 |
Structure Of Positive Allosteric Modulator-Free Active Human Calcium-Sensing Receptor
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2022-01-19 Classification: MEMBRANE PROTEIN Ligands: NAG, TCR, PO4, CA, CLR |
 |
Structure Of Negative Allosteric Modulator-Bound Inactive Human Calcium-Sensing Receptor
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2022-01-19 Classification: MEMBRANE PROTEIN Ligands: YP1 |
 |
Organism: Severe acute respiratory syndrome coronavirus 2
Method: ELECTRON MICROSCOPY Release Date: 2020-12-09 Classification: VIRAL PROTEIN Ligands: U5P, PO4 |
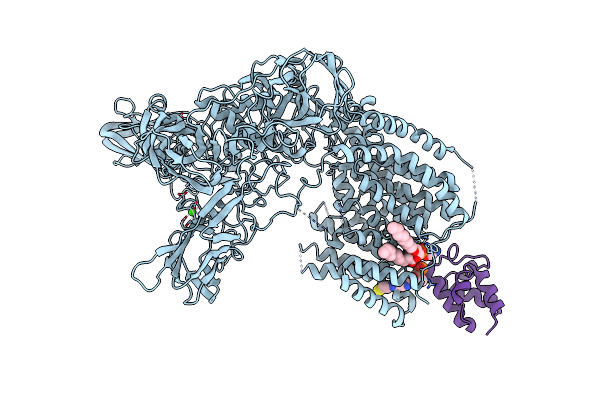 |
Single-Particle Cryo-Em Structure Of Arabinofuranosyltransferase Aftd From Mycobacteria
Organism: Mycobacteroides abscessus subsp. abscessus, Escherichia coli (strain k12)
Method: ELECTRON MICROSCOPY Release Date: 2020-05-13 Classification: MEMBRANE PROTEIN Ligands: CA, 6OU, PNS |
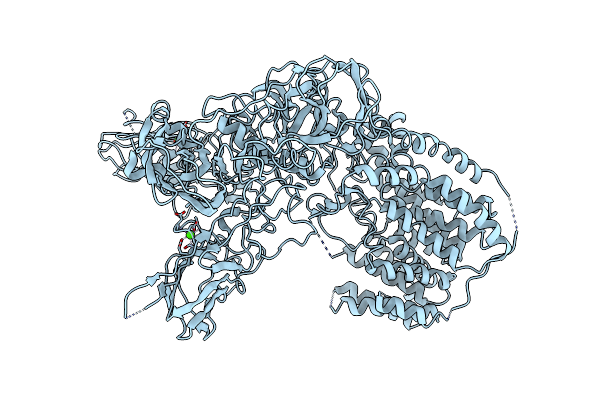 |
Single-Particle Cryo-Em Structure Of Arabinofuranosyltransferase Aftd From Mycobacteria, Mutant R1389S Class 1
Organism: Mycobacteroides abscessus
Method: ELECTRON MICROSCOPY Release Date: 2020-05-13 Classification: MEMBRANE PROTEIN Ligands: CA |
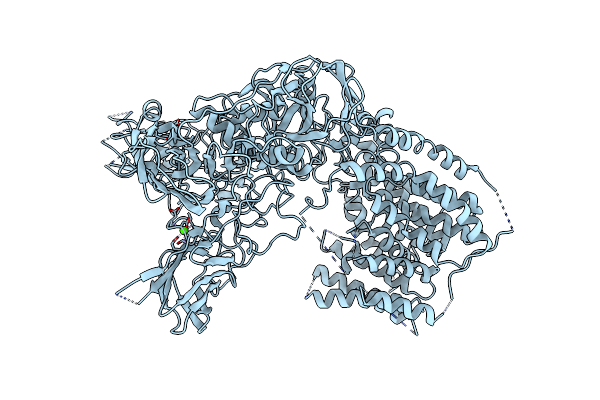 |
Single-Particle Cryo-Em Structure Of Arabinofuranosyltransferase Aftd From Mycobacteria, Mutant R1389S Class 2
Organism: Mycobacteroides abscessus
Method: ELECTRON MICROSCOPY Release Date: 2020-05-13 Classification: MEMBRANE PROTEIN Ligands: CA |

