Search Count: 26
 |
Structure Of The Retron Ia Complex With Hnh Nuclease In The "Down" Orientation
Organism: Escherichia coli
Method: ELECTRON MICROSCOPY Release Date: 2025-04-23 Classification: Transferase/DNA/RNA Ligands: ATP, ZN |
 |
Structure Of The Retron Ia Complex With Hnh Nuclease In The "Up" Orientation
Organism: Escherichia coli
Method: ELECTRON MICROSCOPY Release Date: 2025-04-23 Classification: Transferase/DNA/RNA Ligands: ATP, ZN |
 |
Organism: Escherichia coli
Method: ELECTRON MICROSCOPY Release Date: 2025-04-23 Classification: Transferase/DNA/RNA Ligands: ATP |
 |
Organism: Shewanella oneidensis
Method: X-RAY DIFFRACTION Resolution:3.30 Å Release Date: 2025-01-15 Classification: ELECTRON TRANSPORT Ligands: HEC, CA, PE3 |
 |
Organism: Halorhodospira halophila
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2025-01-15 Classification: SIGNALING PROTEIN Ligands: HC4 |
 |
Stigmatella Aurantica Bacteriophytochrome Protein 2 (Sabphp2), Photosensory Core Module, Investigated At Esrf(Ebs) Id29. Dark Structure.
Organism: Stigmatella aurantiaca
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2025-01-15 Classification: SIGNALING PROTEIN Ligands: BLA, BEN |
 |
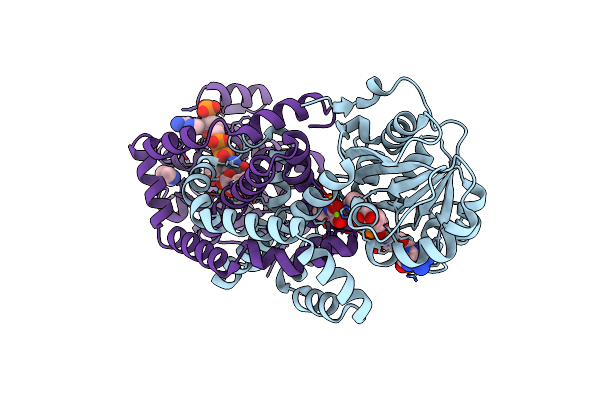
Apo Form Cryo-Em Structure Of Campylobacter Jejune Ketol-Acid Reductoisommerase Crosslinked By Glutaraldehyde
Organism: Campylobacter jejuni
Method: ELECTRON MICROSCOPY Release Date: 2023-02-01 Classification: ISOMERASE Ligands: PTD |
 |
Crystal Structure Of Staphylococcus Aureus Ketol-Acid Reductoisomerase With Hydroxyoxamate Inhibitor 1
Organism: Staphylococcus aureus (strain bovine rf122 / et3-1)
Method: X-RAY DIFFRACTION Resolution:1.67 Å Release Date: 2019-01-30 Classification: OXIDOREDUCTASE Ligands: NDP, MG, EN4, 81B, IMD |
 |
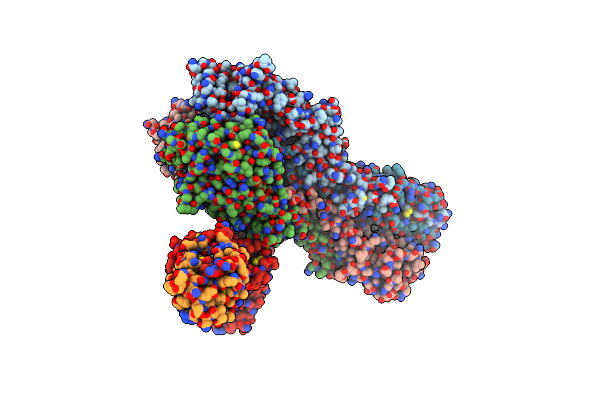
The Crystal Structure Of Neuramindase From A/Canine/Il/11613/2015 (H3N2) Influenza Virus.
Organism: Unidentified influenza virus
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2019-01-30 Classification: VIRAL PROTEIN Ligands: CA, NAG |
 |
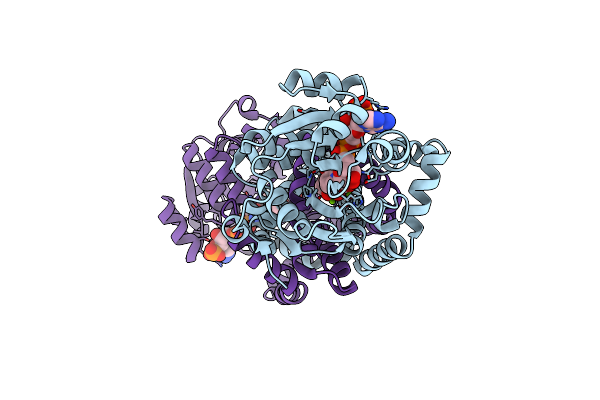
The Crystal Structure Of Hemagglutinin From A/Canine/Il/11613/2015 (H3N2) Influenza Virus.
Organism: Unidentified influenza virus
Method: X-RAY DIFFRACTION Release Date: 2019-01-30 Classification: VIRAL PROTEIN |
 |
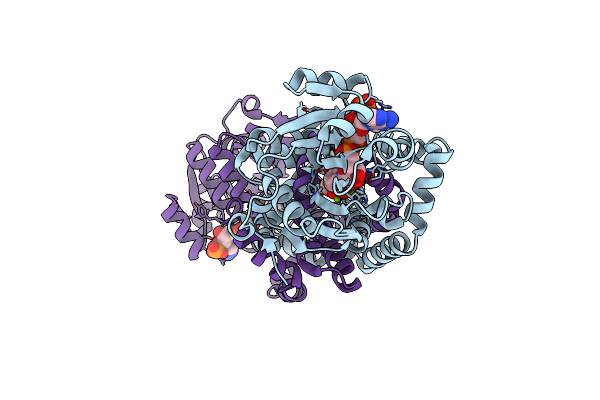
Crystal Structure Of Staphylococcus Aureus Ketol-Acid Reductosimerrase With Hydroxyoxamate Inhibitor 3
Organism: Staphylococcus aureus (strain bovine rf122 / et3-1)
Method: X-RAY DIFFRACTION Resolution:2.09 Å Release Date: 2019-01-16 Classification: OXIDOREDUCTASE Ligands: MG, NAP, EKD, EKA |
 |
Crystal Structure Of Staphylococcus Aureus Ketol-Acid Reductoisomerase With Hydroxyoxamate Inhibitor 2
Organism: Staphylococcus aureus (strain bovine rf122 / et3-1)
Method: X-RAY DIFFRACTION Resolution:1.88 Å Release Date: 2018-12-12 Classification: Oxidoreductase/Inhibitor Ligands: MG, NAP, E9G, E9J |
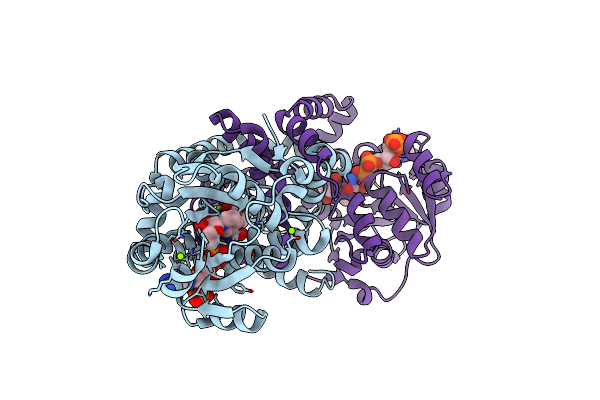 |
Crystal Structure Of Staphylococcus Aureus Ketol-Acid Reductoisomerase In Complex Nadph, Mg2+ And Cpd
Organism: Staphylococcus aureus
Method: X-RAY DIFFRACTION Resolution:1.59 Å Release Date: 2017-10-25 Classification: ISOMERASE Ligands: 9TY, MG, NDP |
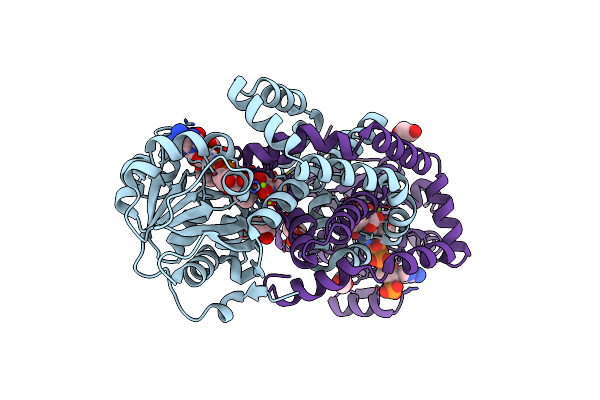 |
Crystal Structures Of Staphylococcus Aureus Ketol-Acid Reductoisomerase In Complex With Two Transition State Analogs That Have Biocidal Activity.
Organism: Staphylococcus aureus
Method: X-RAY DIFFRACTION Resolution:1.37 Å Release Date: 2017-10-18 Classification: ISOMERASE Ligands: MG, 40E, GOL, NDP, HIO, EDO |
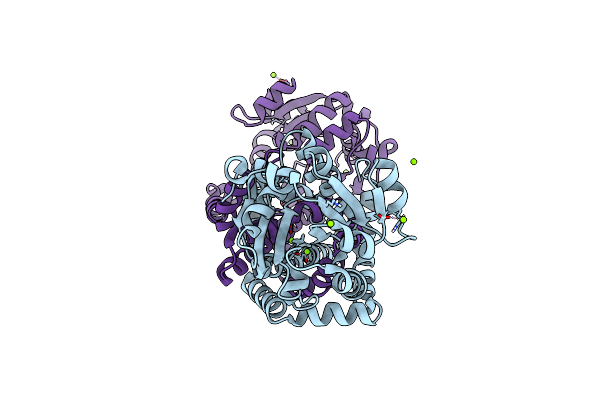 |
Crystal Structure Of Mycobacterium Tuberculosis Ketol-Acid Reductoisomerase In Complex With Mg2+
Organism: Mycobacterium tuberculosis
Method: X-RAY DIFFRACTION Resolution:1.00 Å Release Date: 2016-02-17 Classification: OXIDOREDUCTASE Ligands: MG, CL, NA |
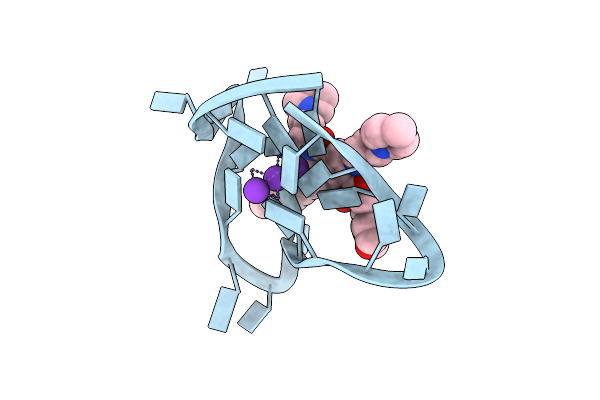 |
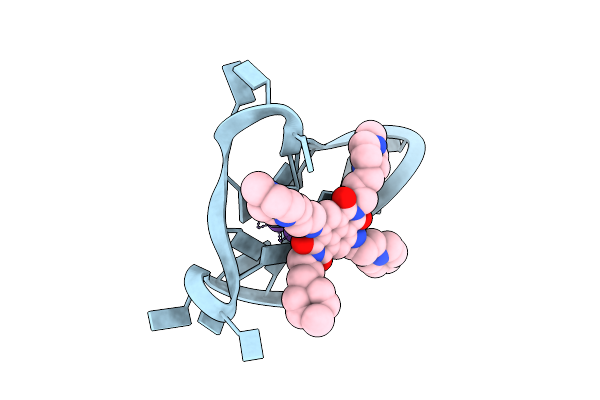
Crystal Structure Of An Intramolecular Human Telomeric Dna G-Quadruplex 21-Mer Bound By The Naphthalene Diimide Compound Mm41.
Organism: Synthetic dna
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2013-01-30 Classification: DNA Ligands: K, 0DX |
 |
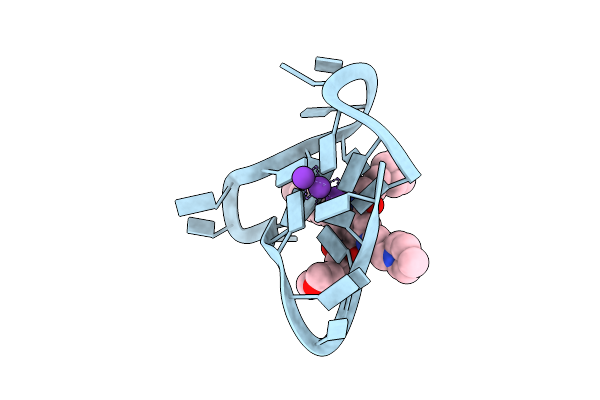
Crystal Structure Of An Intramolecular Human Telomeric Dna G-Quadruplex 21-Mer Bound By The Naphthalene Diimide Compound Bmsg-Sh-3
Organism: Synthetic dna
Method: X-RAY DIFFRACTION Resolution:2.75 Å Release Date: 2013-01-16 Classification: DNA Ligands: K, R8G |
 |
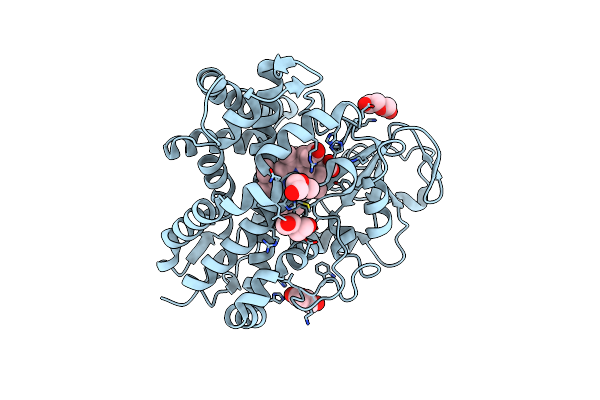
Crystal Structure Of An Intramolecular Human Telomeric Dna G-Quadruplex Bound By The Naphthalene Diimide Compound, Mm41
Organism: Synthetic dna
Method: X-RAY DIFFRACTION Resolution:1.95 Å Release Date: 2013-01-02 Classification: DNA Ligands: K, 0DX |
 |
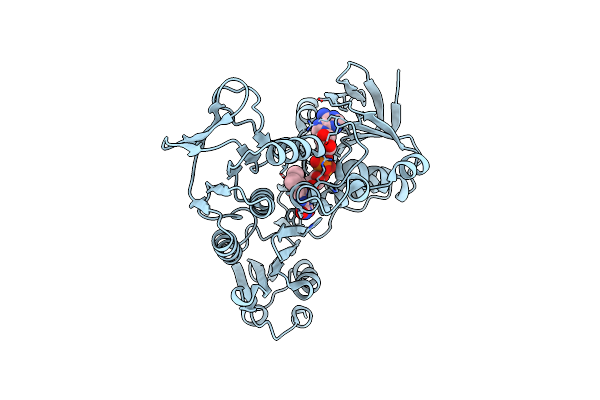
Organism: Novosphingobium aromaticivorans
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2012-08-15 Classification: OXIDOREDUCTASE Ligands: HEM, PEG |
 |
Crystal Structure Of Ferredoxin Reductase Arr From Novosphingobium Aromaticivorans
Organism: Novosphingobium aromaticivorans
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2010-06-23 Classification: OXIDOREDUCTASE Ligands: FAD |

