Search Count: 13
 |
Organism: Rhizomucor miehei
Method: X-RAY DIFFRACTION Resolution:1.49 Å Release Date: 2019-03-13 Classification: HYDROLASE Ligands: NAG, EDO |
 |
Organism: Rhizomucor miehei
Method: X-RAY DIFFRACTION Resolution:1.45 Å Release Date: 2019-03-13 Classification: HYDROLASE Ligands: NAG |
 |
A Naturally Variable Residue In The S1 Subsite Of M1-Family Aminopeptidases Modulates Catalytic Properties And Promotes Functional Specialization
Organism: Plasmodium falciparum
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2013-08-07 Classification: HYDROLASE Ligands: ARG, ZN, MG |
 |
Co-Crystal Structure Of Wild Type Rat Polymerase Beta: Enzyme-Dna Binary Complex
Organism: Rattus norvegicus
Method: X-RAY DIFFRACTION Resolution:2.25 Å Release Date: 2013-01-16 Classification: Transferase/DNA |
 |
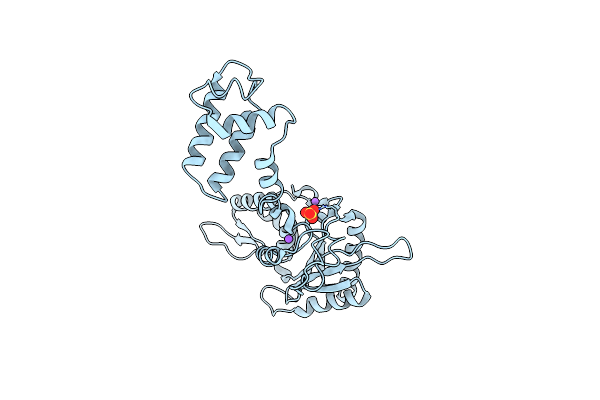
Co-Crystal Structure Of K72E Variant Of Rat Polymerase Beta: Enzyme-Dna Binary Complex
Organism: Rattus norvegicus
Method: X-RAY DIFFRACTION Resolution:2.27 Å Release Date: 2013-01-16 Classification: Transferase/DNA |
 |
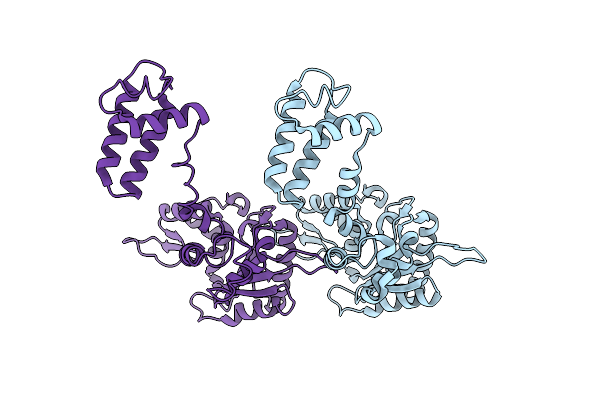
Organism: Rattus norvegicus
Method: X-RAY DIFFRACTION Resolution:2.66 Å Release Date: 2013-01-16 Classification: TRANSFERASE |
 |
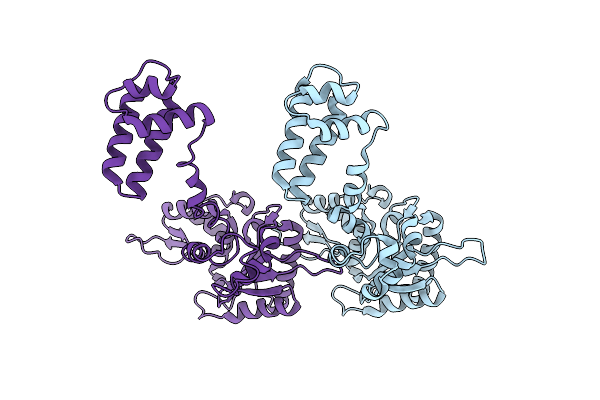
Organism: Rattus norvegicus
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2012-12-05 Classification: TRANSFERASE |
 |
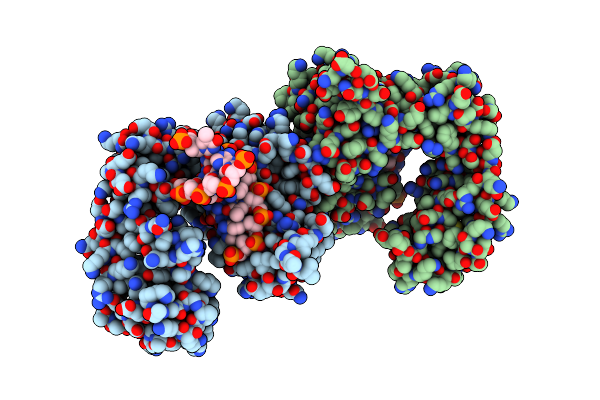
Organism: Rattus norvegicus
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2012-12-05 Classification: TRANSFERASE, LYASE |
 |
Co-Crystal Structure Of Rat Dna Polymerase Beta Mutator I260Q: Enzyme-Dna-Ddttp
Organism: Rattus norvegicus
Method: X-RAY DIFFRACTION Resolution:2.72 Å Release Date: 2012-12-05 Classification: TRANSFERASE, LYASE/DNA Ligands: D3T, NA |
 |
A Bestatin-Based Chemical Biology Strategy Reveals Distinct Roles For Malaria M1- And M17-Family Aminopeptidases
Organism: Plasmodium falciparum
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2011-09-28 Classification: HYDROLASE/HYDROLASE INHIBITOR Ligands: ZN, MG, BTJ |
 |
A Bestatin-Based Chemical Biology Strategy Reveals Distinct Roles For Malaria M1- And M17-Family Aminopeptidases
Organism: Plasmodium falciparum
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2011-09-28 Classification: HYDROLASE/HYDROLASE INHIBITOR Ligands: CO3, ZN, DGZ, SO4, 1PE, 2PE |
 |
Organism: Plasmodium falciparum fcb1/columbia
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2011-03-16 Classification: HYDROLASE/HYDROLASE INHIBITOR Ligands: ZN, MG, GOL, D66 |
 |
Organism: Plasmodium falciparum fcb1/columbia
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2011-03-16 Classification: HYDROLASE/HYDROLASE INHIBITOR Ligands: ZN, D50, GOL, MG |

