Search Count: 174
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-12-10 Classification: MEMBRANE PROTEIN |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-12-10 Classification: MEMBRANE PROTEIN Ligands: UKX |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-12-10 Classification: MEMBRANE PROTEIN Ligands: A1CCA |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-12-10 Classification: MEMBRANE PROTEIN Ligands: A1CCA, A1B1I |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-12-10 Classification: MEMBRANE PROTEIN Ligands: X3U |
 |
Cryo-Em Structure Of Human Sv2A In Complex With Levetiracetam And Ucb1244283
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-12-10 Classification: MEMBRANE PROTEIN Ligands: UKX, A1B1J |
 |
An Antibiotic Biosynthesis Monooxygenase Family Protein From Streptomyces Sp. Ma37
Organism: Streptomyces sp. ma37
Method: X-RAY DIFFRACTION Release Date: 2025-11-26 Classification: ANTIBIOTIC |
 |
Crystal Structure Of A Polyketide Abm/Scha-Like Domain-Containing Protein Whie-Orfi From Streptomyces Coelicolor
Organism: Streptomyces coelicolor
Method: X-RAY DIFFRACTION Release Date: 2025-11-26 Classification: ANTIBIOTIC |
 |
Structure Of The Sweet Receptor Bound To Sucralose In The Loose State, Extracellular Domain
Organism: Homo sapiens, Mus musculus
Method: ELECTRON MICROSCOPY Release Date: 2025-11-19 Classification: SIGNALING PROTEIN |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-10-15 Classification: CHAPERONE/ONCOPROTEIN Ligands: BTB, CA, GDP, A1CRY, MG |
 |
Organism: Homo sapiens, Mus musculus
Method: ELECTRON MICROSCOPY Release Date: 2025-09-03 Classification: SIGNALING PROTEIN |
 |
Organism: Homo sapiens, Mus musculus
Method: ELECTRON MICROSCOPY Release Date: 2025-09-03 Classification: SIGNALING PROTEIN |
 |
Organism: Homo sapiens, Mus musculus
Method: ELECTRON MICROSCOPY Release Date: 2025-09-03 Classification: SIGNALING PROTEIN |
 |
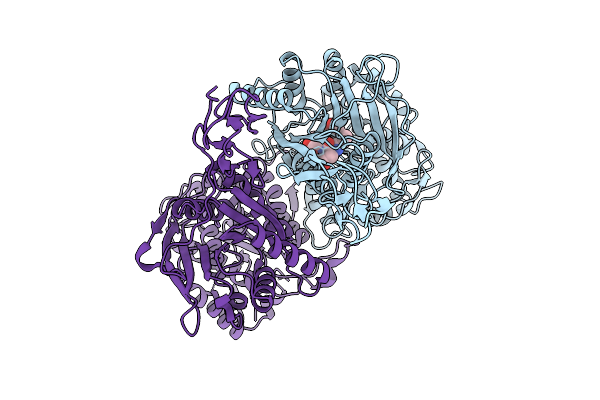
Structure Of The Sweet Receptor Bound To Advantame In The Compact State, Extracellular Domain
Organism: Homo sapiens, Mus musculus
Method: ELECTRON MICROSCOPY Release Date: 2025-09-03 Classification: SIGNALING PROTEIN Ligands: A1CD7 |
 |
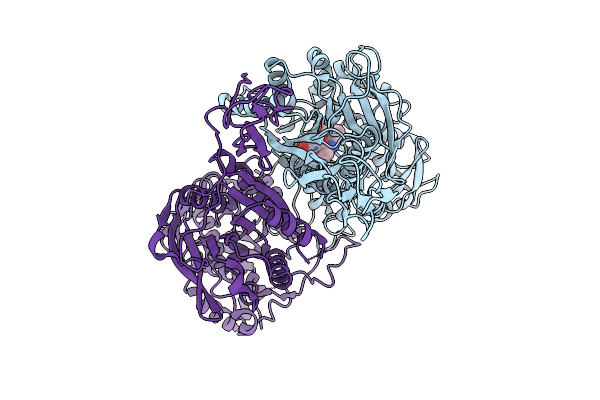
Structure Of The Sweet Receptor Bound To Advantame In The Loose State, Extracellular Domain
Organism: Homo sapiens, Mus musculus
Method: ELECTRON MICROSCOPY Release Date: 2025-09-03 Classification: SIGNALING PROTEIN Ligands: A1CD7 |
 |
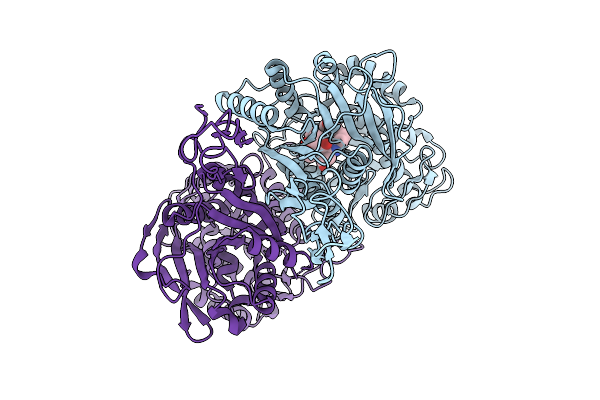
Structure Of The Human Sweet Receptor Bound To Advantame In The Compact State, Extracellular Domain
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-09-03 Classification: SIGNALING PROTEIN Ligands: A1CD7 |
 |
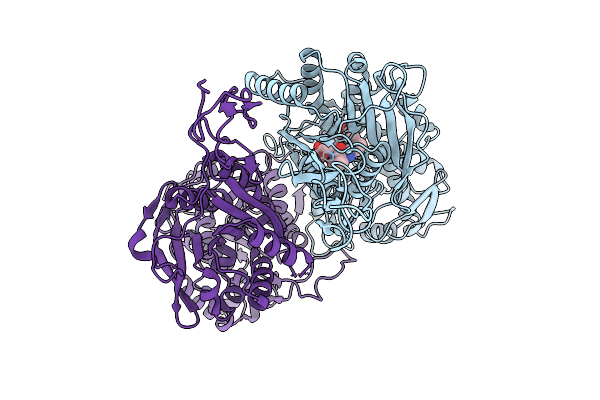
Structure Of The Human Sweet Receptor Bound To Advantame In The Intermediate State, Extracellular Domain
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-09-03 Classification: SIGNALING PROTEIN Ligands: A1CD7 |
 |
Structure Of The Human Sweet Receptor Bound To Advantame In The Loose State, Extracellular Domain
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-09-03 Classification: SIGNALING PROTEIN Ligands: A1CD7 |
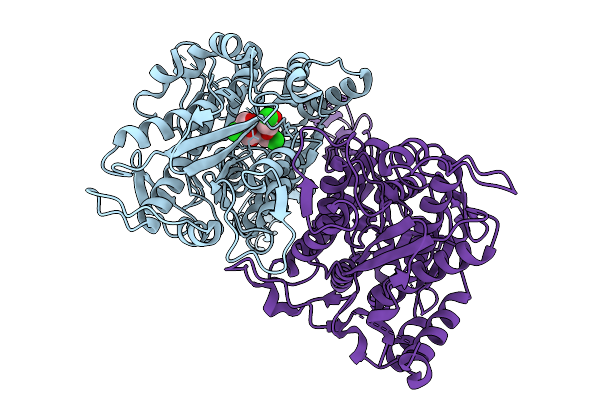 |
Structure Of The Sweet Receptor Bound To Sucralose In The Compact State, Extracellular Domain
Organism: Homo sapiens, Mus musculus
Method: ELECTRON MICROSCOPY Release Date: 2025-08-27 Classification: SIGNALING PROTEIN |
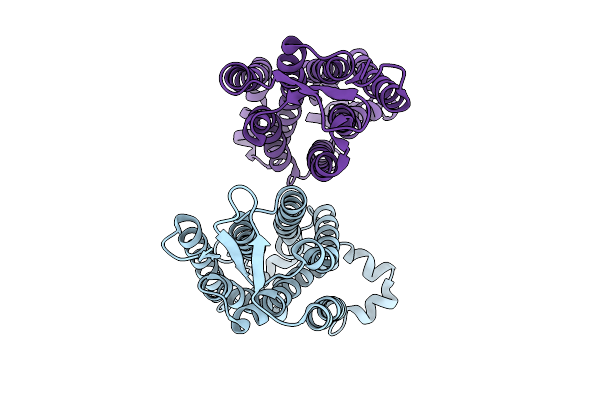 |
Structure Of The Sweet Receptor Bound To Sucralose In The Compact State, Transmembrane Domain
Organism: Homo sapiens, Mus musculus
Method: ELECTRON MICROSCOPY Release Date: 2025-08-27 Classification: SIGNALING PROTEIN |

