Planned Maintenance: Some services may turn out to be unavailable from 15th January, 2026 to 16th January, 2026. We apologize for the inconvenience!
Planned Maintenance: Some services may turn out to be unavailable from 15th January, 2026 to 16th January, 2026. We apologize for the inconvenience!
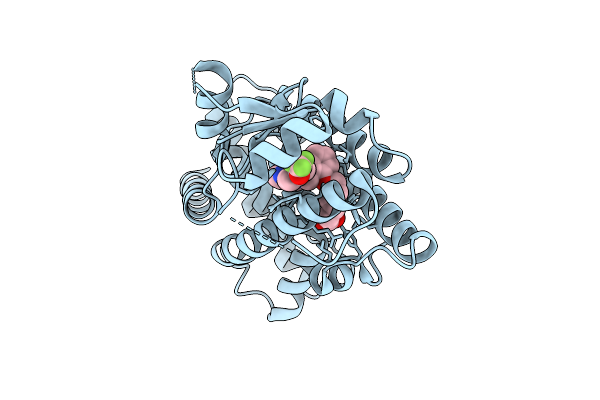 |
X-Ray Structure Of Human Ppar Delta Ligand Binding Domain In Complex With A Synthetic Agonist 16A
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-12-24 Classification: NUCLEAR PROTEIN Ligands: A1D86 |
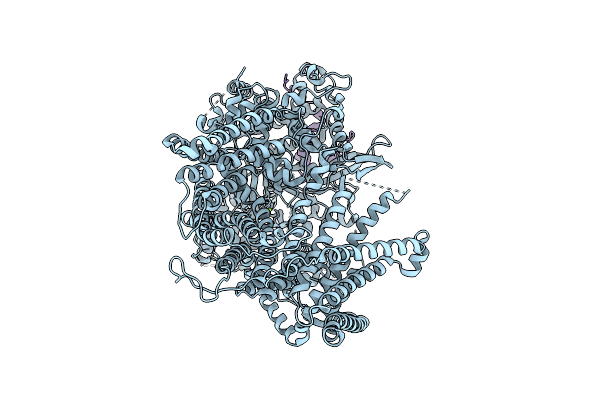 |
Organism: Saccharomyces cerevisiae
Method: ELECTRON MICROSCOPY Release Date: 2025-12-17 Classification: MEMBRANE PROTEIN Ligands: MG |
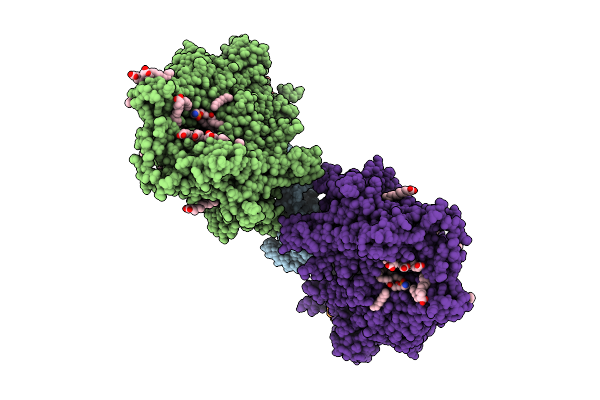 |
Organism: Saccharomyces cerevisiae
Method: ELECTRON MICROSCOPY Release Date: 2025-12-17 Classification: MEMBRANE PROTEIN Ligands: MG, DD9, D10, HP6, XKP, PEF, PLM, DCR |
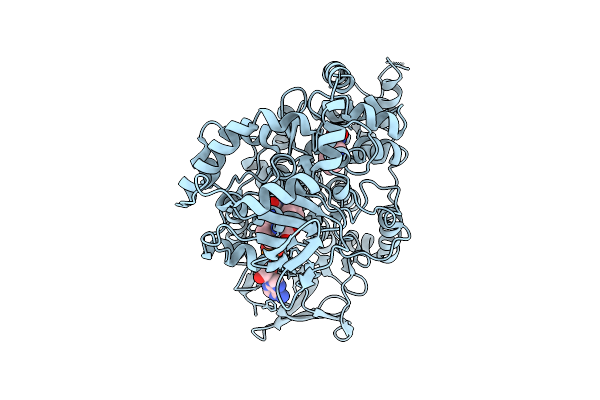 |
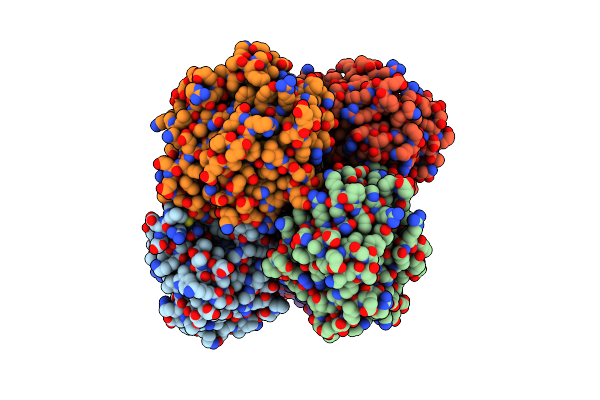
Organism: Aetokthonos hydrillicola thurmond2011
Method: X-RAY DIFFRACTION Release Date: 2025-12-10 Classification: FLAVOPROTEIN Ligands: FAD, TRP |
 |
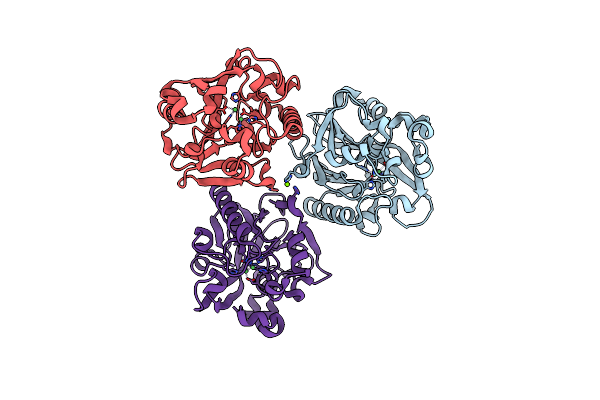
Crystal Structure Of N-Acyl Homoserine Lactonase Ahlx Mutant M41(E77I/D177G/T243Y/H255L)
Organism: Salinicola salarius
Method: X-RAY DIFFRACTION Resolution:2.16 Å Release Date: 2025-04-23 Classification: HYDROLASE Ligands: NI |
 |
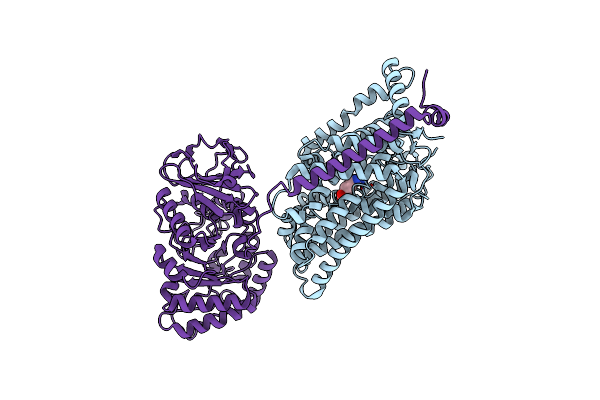
Organism: Salinicola salarius
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2025-04-23 Classification: HYDROLASE Ligands: NI, MG |
 |
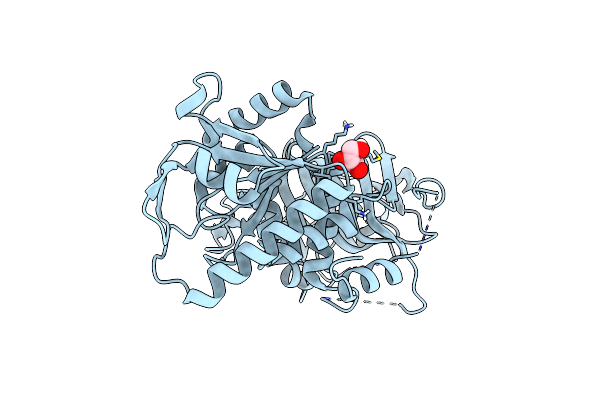
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-04-23 Classification: MEMBRANE PROTEIN Ligands: ARG |
 |
Organism: Escherichia coli o1:k1 / apec
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2025-04-09 Classification: IMMUNE SYSTEM Ligands: GOL, CL |
 |
Organism: Escherichia coli o1:k1 / apec
Method: X-RAY DIFFRACTION Resolution:2.04 Å Release Date: 2025-04-09 Classification: IMMUNE SYSTEM Ligands: GOL, CL |
 |
Organism: Escherichia coli o1:k1 / apec
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2025-04-09 Classification: IMMUNE SYSTEM Ligands: PRP, MES, GOL, MG, SO4 |
 |
Organism: Escherichia coli o1:k1 / apec
Method: X-RAY DIFFRACTION Resolution:1.72 Å Release Date: 2025-04-09 Classification: IMMUNE SYSTEM Ligands: PO4 |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-04-02 Classification: MEMBRANE PROTEIN |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-04-02 Classification: MEMBRANE PROTEIN Ligands: ARG |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-03-12 Classification: MEMBRANE PROTEIN Ligands: ARG |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.95 Å Release Date: 2025-02-26 Classification: IMMUNOSUPPRESSANT/INHIBITOR Ligands: A1LV3 |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-01-22 Classification: MEMBRANE PROTEIN |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-01-22 Classification: MEMBRANE PROTEIN Ligands: LYS |
 |
Cryo-Em Structure And Rational Engineering Of A Novel Efficient Ochratoxin A-Detoxifying Amidohydrolase
Organism: Pseudoxanthomonas wuyuanensis
Method: ELECTRON MICROSCOPY Release Date: 2024-12-18 Classification: HYDROLASE Ligands: ZN, 97U |
 |
Cryo-Electron Microscopic Structure Of An Amide Hydrolase From Pseudoxanthomonas Wuyuanensis
Organism: Pseudoxanthomonas wuyuanensis
Method: ELECTRON MICROSCOPY Release Date: 2024-10-09 Classification: HYDROLASE Ligands: ZN |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2024-09-25 Classification: TRANSPORT PROTEIN/HYDROLASE Ligands: NAG, TGI |