Search Count: 12
 |
1,3-Alpha-1,4-Beta-D-Galactose-4-Sulfate-3,6-Anhydro-D-Galactose 4 Galactohydrolase
Organism: Pseudoalteromonas carrageenovora
Method: X-RAY DIFFRACTION Resolution:1.54 Å Release Date: 2001-01-16 Classification: HYDROLASE Ligands: CD, CL |
 |
Organism: Pseudomonas aeruginosa
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2001-01-12 Classification: TRANSFERASE Ligands: SO4 |
 |
Organism: Bacillus cereus
Method: X-RAY DIFFRACTION Resolution:1.85 Å Release Date: 2000-08-25 Classification: HYDROLASE Ligands: ZN, BCT |
 |
Organism: Streptococcus pneumoniae
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2000-05-25 Classification: PENICILLIN-BINDING PROTEIN Ligands: SO4 |
 |
Organism: Streptococcus pneumoniae
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2000-05-25 Classification: CELL CYCLE Ligands: CES, KEF |
 |
Organism: Bacillus cereus
Method: X-RAY DIFFRACTION Resolution:1.85 Å Release Date: 1998-09-23 Classification: HYDROLASE Ligands: ZN, BCT |
 |
Organism: Bacteroides fragilis
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 1998-09-23 Classification: HYDROLASE Ligands: ZN, NA |
 |
Organism: Bacillus cereus
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 1996-08-28 Classification: HYDROLASE (ACTING IN CYCLIC AMIDES) Ligands: ZN |
 |
Comparison Of The Structures Of Wild Type And A N313T Mutant Of Escherichia Coli Glyceraldehyde 3-Phosphate Dehydrogenases: Implication For Nad Binding And Cooperativity
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 1996-03-08 Classification: OXIDOREDUCTASE (ALDEHYDE(D)-NAD+(A)) Ligands: NAD |
 |
Comparison Of The Structures Of Wild Type And A N313T Mutant Of Escherichia Coli Glyceraldehyde 3-Phosphate Dehydrogenases: Implication For Nad Binding And Cooperativity
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.17 Å Release Date: 1996-03-08 Classification: OXIDOREDUCTASE (ALDEHYDE(D)-NAD+(A)) Ligands: NAD |
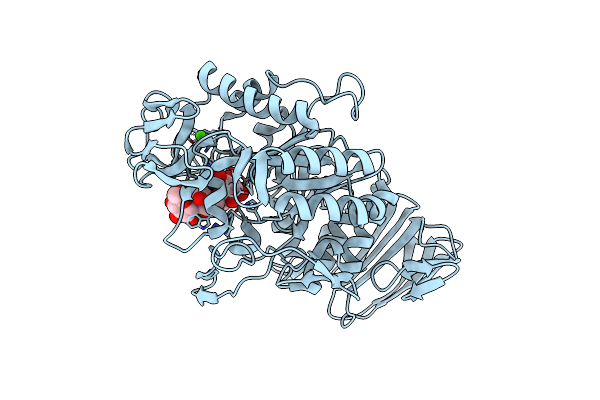 |
The Active Center Of A Mammalian Alpha-Amylase. The Structure Of The Complex Of A Pancreatic Alpha-Amylase With A Carbohydrate Inhibitor Refined To 2.2 Angstroms Resolution
Organism: Sus scrofa
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 1995-05-24 Classification: HYDROLASE (O-GLYCOSYL) Ligands: CL, CA |
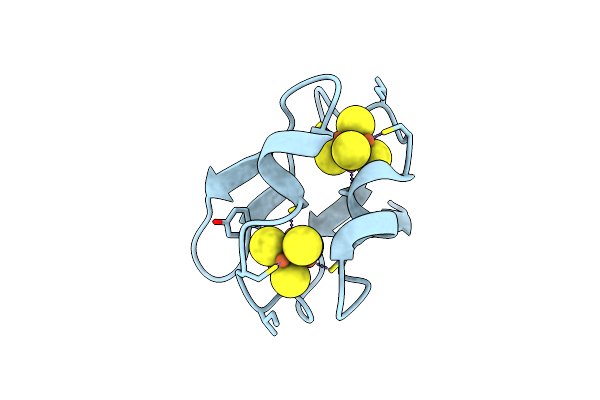 |
Refined Crystal Structure Of The 2[4Fe-4S] Ferredoxin From Clostridium Acidurici At 1.84 Angstroms Resolution
Organism: Clostridium acidurici
Method: X-RAY DIFFRACTION Resolution:1.84 Å Release Date: 1994-08-31 Classification: ELECTRON TRANSPORT Ligands: SF4 |

