Search Count: 298
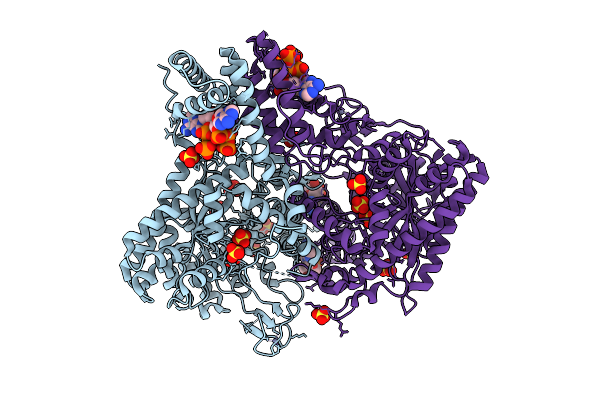 |
Crystal Structure Of S. Thermophilus Class Iii Ribonucleotide Reductase Bound To Datp
Organism: Streptococcus thermophilus
Method: X-RAY DIFFRACTION Release Date: 2025-12-17 Classification: OXIDOREDUCTASE Ligands: MG, DTP, ZN, SO4 |
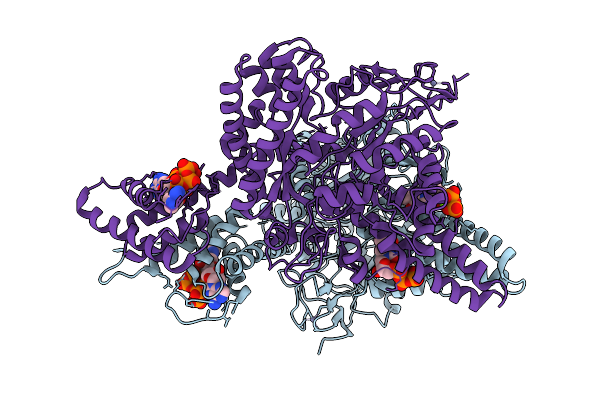 |
Organism: Streptococcus thermophilus
Method: ELECTRON MICROSCOPY Release Date: 2025-12-17 Classification: OXIDOREDUCTASE Ligands: TTP, DTP, MG, ZN |
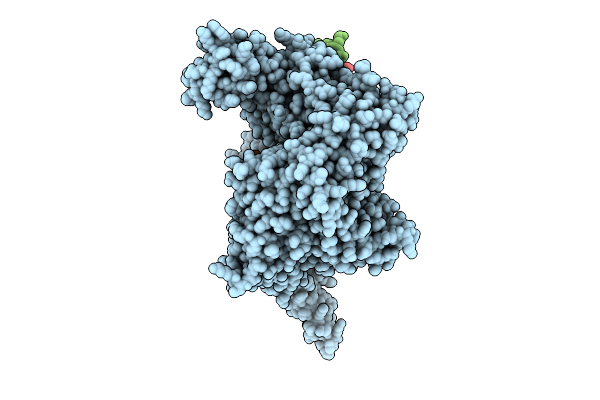 |
Structure Of Human Line-1 Orf2P With Endogenous Dna And Rna/Cdna Hybrid Bound To Dntp And Mn2+
Organism: Homo sapiens, Trichoplusia ni
Method: ELECTRON MICROSCOPY Release Date: 2025-10-22 Classification: GENE REGULATION Ligands: DTP, ZN, MN |
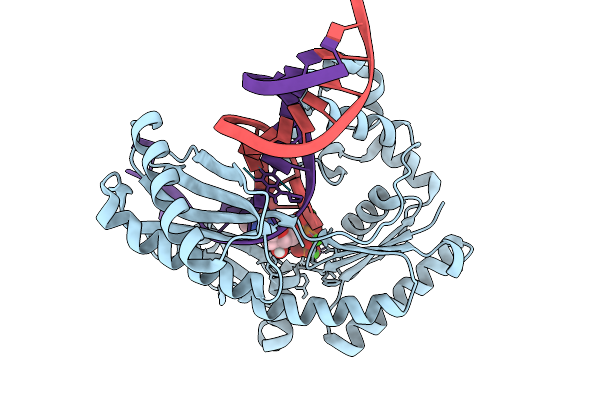 |
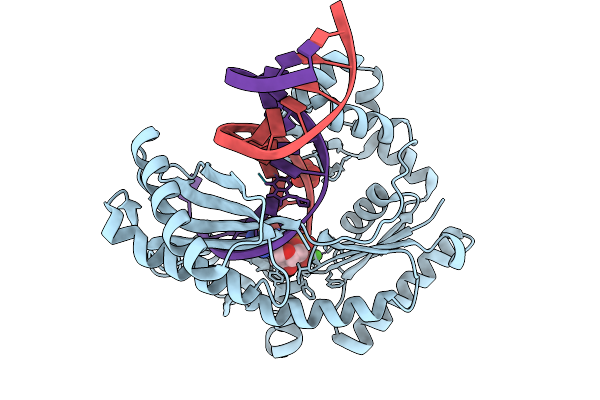
Dpo4 Dna Polymerase (Wild Type) In Complex With Dna Containing An 8Oxog Template Lesion
Organism: Saccharolobus solfataricus p2, Synthetic construct
Method: X-RAY DIFFRACTION Release Date: 2025-09-24 Classification: TRANSFERASE/DNA Ligands: CA, DTP |
 |
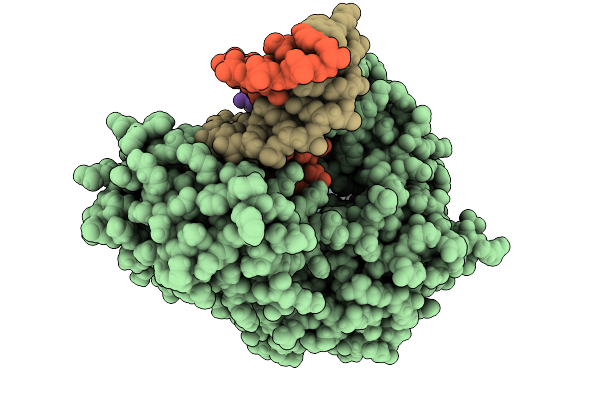
Dpo4 Dna Polymerase (R332A) In Complex With Dna Containing An 8Oxog Template Lesion
Organism: Saccharolobus solfataricus, Synthetic construct
Method: X-RAY DIFFRACTION Release Date: 2025-09-24 Classification: TRANSFERASE/DNA Ligands: DTP, CA, PO4 |
 |
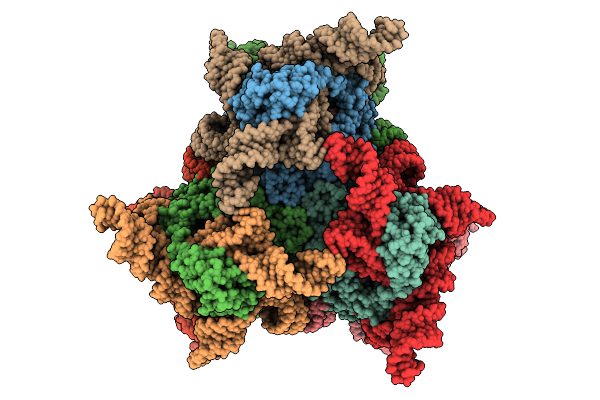
Dpo4 Dna Polymerase (R336A) In Complex With Dna Containing An 8Oxog Template Lesion
Organism: Saccharolobus solfataricus, Synthetic construct
Method: X-RAY DIFFRACTION Release Date: 2025-09-24 Classification: TRANSFERASE/DNA Ligands: DTP, CA, PO4 |
 |
Organism: Escherichia coli
Method: ELECTRON MICROSCOPY Release Date: 2025-08-20 Classification: ANTIVIRAL PROTEIN/RNA/DNA Ligands: DTP, MG |
 |
Cryo-Em Structure Of The Strand Displacement Complex (Iv) Of Yeast Mitochondrial Dna Polymerase Gamma (Mip1) With Downstream Dna
Organism: Saccharomyces cerevisiae, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2025-03-19 Classification: Transferase/DNA Ligands: DTP, MG |
 |
Cryo-Em Structure Of The Strand Displacement Complex (I) Of Yeast Mitochondrial Dna Polymerase Gamma (Mip1) With Downstream Dna
Organism: Saccharomyces cerevisiae, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2025-03-19 Classification: Transferase/DNA Ligands: MG, DTP, 2DT |
 |
Cryo-Em Structure Of The Strand Displacement Complex (Ii) Of Yeast Mitochondrial Dna Polymerase Gamma (Mip1) With Downstream Dna
Organism: Saccharomyces cerevisiae, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2025-03-19 Classification: Transferase/DNA Ligands: DTP, MG, 2DT |
 |
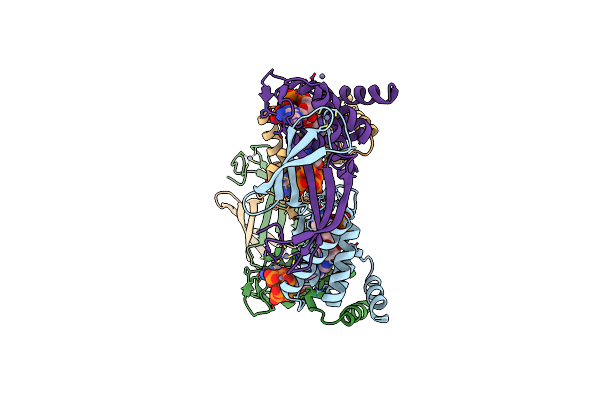
Cryo-Em Structure Of The Strand Displacement Complex (Iii) Of Yeast Mitochondrial Dna Polymerase Gamma (Mip1) With Downstream Dna
Organism: Saccharomyces cerevisiae, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2025-03-19 Classification: REPLICATION Ligands: DTP, MG, 2DT |
 |
Transcription Repressor Nrdr From E. Coli, Atp/Datp-Bound State, Semet Protein
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Release Date: 2025-03-19 Classification: DNA BINDING PROTEIN Ligands: ATP, DTP, ZN |
 |
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Release Date: 2025-03-19 Classification: DNA BINDING PROTEIN Ligands: DTP, ANP, ZN, MG |
 |
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Release Date: 2025-03-19 Classification: DNA BINDING PROTEIN Ligands: DTP, ADP, ZN, MG |
 |
Crystal Structure Of The Deinococcus Wulumuqiensis Cd-Ntase Dwcdnb In Complex With Datp
Organism: Deinococcus wulumuqiensis
Method: X-RAY DIFFRACTION Resolution:2.19 Å Release Date: 2025-01-29 Classification: TRANSFERASE Ligands: DTP, MG |
 |
Organism: Human immunodeficiency virus 1, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2024-12-11 Classification: TRANSCRIPTION, TRANSFERASE/DNA Ligands: MG, DTP |
 |
Organism: Human immunodeficiency virus 1, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2024-12-11 Classification: TRANSCRIPTION, TRANSFERASE/DNA Ligands: DTP, MG |
 |
Organism: Human immunodeficiency virus 1, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2024-12-11 Classification: TRANSCRIPTION, TRANSFERASE/DNA Ligands: MG, DTP |
 |
Organism: Human immunodeficiency virus 1, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2024-12-11 Classification: TRANSCRIPTION, TRANSFERASE/DNA Ligands: MG, DTP |
 |
Organism: Human immunodeficiency virus 1, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2024-12-11 Classification: TRANSCRIPTION, TRANSFERASE/DNA Ligands: MG, DTP |

