Search Count: 196
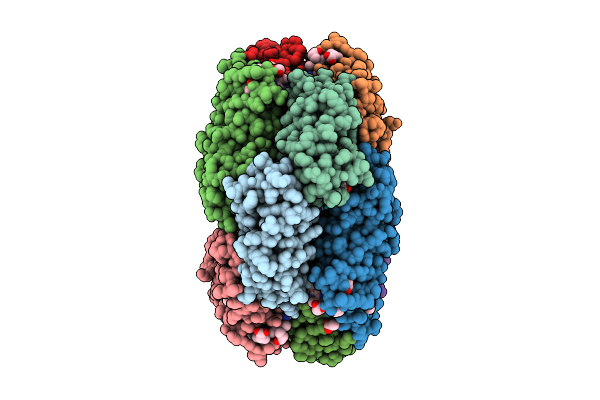 |
X-Ray Structure Of The Adduct Formed By Dirhodium Tetraacetate With A C-Phycocyanin
Organism: Galdieria phlegrea
Method: X-RAY DIFFRACTION Release Date: 2025-12-17 Classification: PHOTOSYNTHESIS Ligands: CYC, A1JTN, ACT, DOD |
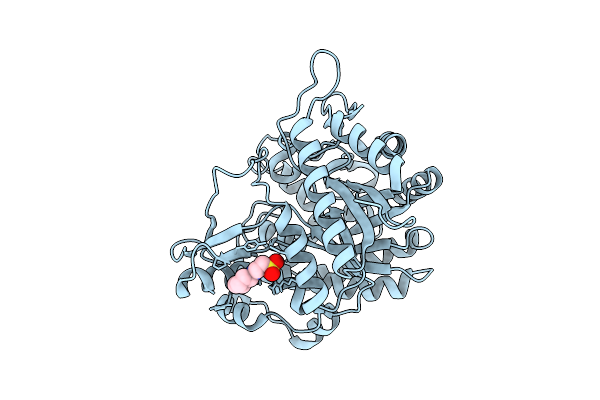 |
Neutron Structure Of Gh1 Beta-Glucosidase Td2F2 Ligand-Free Form At Room Temperature
Organism: Metagenome
Method: X-RAY DIFFRACTION, NEUTRON DIFFRACTION Release Date: 2025-10-29 Classification: HYDROLASE Ligands: NHE, DOD |
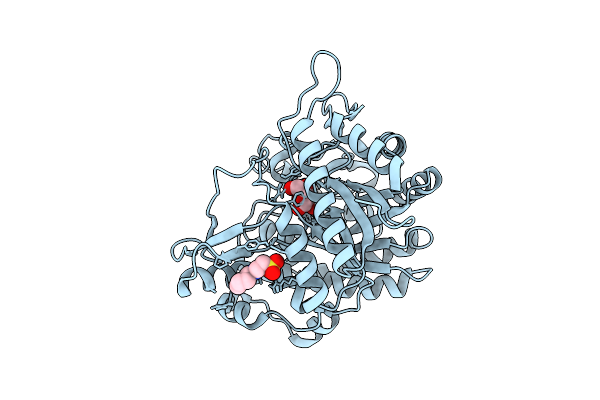 |
Neutron Structure Of Gh1 Beta-Glucosidase Td2F2 Glucose Complex At Room Temperature
Organism: Metagenome
Method: X-RAY DIFFRACTION, NEUTRON DIFFRACTION Release Date: 2025-10-29 Classification: HYDROLASE Ligands: BGC, NHE, DOD |
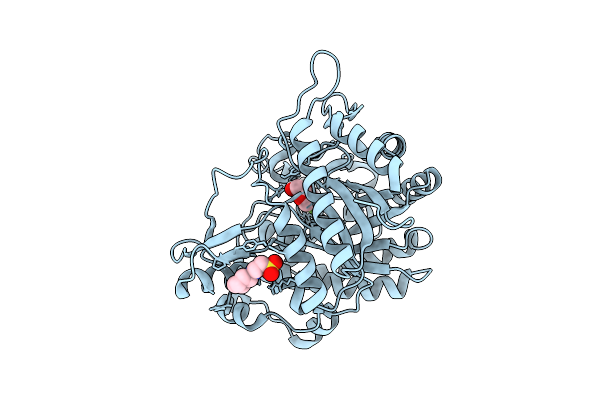 |
Neutron Structure Of Gh1 Beta-Glucosidase Td2F2 2F-Glc Complex At Room Temperature
Organism: Metagenome
Method: X-RAY DIFFRACTION, NEUTRON DIFFRACTION Release Date: 2025-10-29 Classification: HYDROLASE Ligands: G2F, NHE, DOD |
 |
Neutron And X-Ray Joint Refined Structure Of A Copper-Containing Nitrite Reductase (C135A Mutant) In Complex With Nitrite
Organism: Geobacillus thermodenitrificans ng80-2
Method: X-RAY DIFFRACTION, NEUTRON DIFFRACTION Release Date: 2025-06-11 Classification: OXIDOREDUCTASE Ligands: MPD, MRD, NO2, CU, DOD |
 |
Organism: Azoarcus sp. cib
Method: X-RAY DIFFRACTION Release Date: 2025-03-05 Classification: OXIDOREDUCTASE Ligands: BYC, GOL, SF4, BJ8, TRS, DOD |
 |
Organism: Azoarcus sp. cib
Method: X-RAY DIFFRACTION Release Date: 2025-03-05 Classification: OXIDOREDUCTASE Ligands: BYC, GOL, SF4, BJ8, TRS, DOD |
 |
Neutron Structure Of Hydrogenated Hen Egg-White Lysozyme At Room Temperature
Organism: Gallus gallus
Method: NEUTRON DIFFRACTION Resolution:1.10 Å Release Date: 2024-11-06 Classification: HYDROLASE Ligands: NO3, ACT, DOD |
 |
Neutron Structure Of Perdeuterated Hen Egg-White Lysozyme At Room Temperature
Organism: Gallus gallus
Method: NEUTRON DIFFRACTION Resolution:0.91 Å Release Date: 2024-11-06 Classification: HYDROLASE Ligands: NO3, ACT, DOD |
 |
Joint X-Ray/Neutron Structure Of Thermus Thermophilus Serine Hydroxymethyltransferase (Tthshmt) In Internal Aldimine State And Folinic Acid Bound
Organism: Thermus thermophilus hb8
Method: X-RAY DIFFRACTION, NEUTRON DIFFRACTION Resolution:2.30 Å, 2.00 Å Release Date: 2024-08-28 Classification: TRANSFERASE Ligands: SO4, FFO, ACT, DOD |
 |
Organism: Alcaligenes faecalis
Method: X-RAY DIFFRACTION, NEUTRON DIFFRACTION Resolution:1.50 Å, 1.90 Å Release Date: 2024-02-14 Classification: ELECTRON TRANSPORT Ligands: CU, SO4, DOD |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION, NEUTRON DIFFRACTION Resolution:1.190 Å, 1.900 Å Release Date: 2023-11-22 Classification: TRANSPORT PROTEIN Ligands: PIT, DOD |
 |
Joint X-Ray/Neutron Structure Of Thermus Thermophilus Serine Hydroxymethyltransferase (Tthshmt) In Internal Aldimine State With L-Ser Bound In A Pre-Michalis Complex
Organism: Thermus thermophilus hb8
Method: X-RAY DIFFRACTION, NEUTRON DIFFRACTION Resolution:2.30 Å, 2.00 Å Release Date: 2023-08-16 Classification: TRANSFERASE Ligands: SO4, SER, DOD |
 |
Joint X-Ray/Neutron Structure Of Thermus Thermophilus Serine Hydroxymethyltransferase (Tthshmt) In Internal Aldimine State
Organism: Thermus thermophilus hb8
Method: X-RAY DIFFRACTION, NEUTRON DIFFRACTION Resolution:2.30 Å, 2.00 Å Release Date: 2023-08-16 Classification: TRANSFERASE Ligands: SO4, DOD |
 |
Joint X-Ray/Neutron Structure Of Aspastate Aminotransferase (Aat) In Complex With Pyridoxamine 5'-Phosphate (Pmp)
Organism: Sus scrofa
Method: X-RAY DIFFRACTION, NEUTRON DIFFRACTION Resolution:1.70 Å, 2.22 Å Release Date: 2022-09-28 Classification: TRANSFERASE Ligands: PLA, PMP, DOD |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION, NEUTRON DIFFRACTION Resolution:1.00 Å, 2.25 Å Release Date: 2022-07-20 Classification: RNA Ligands: SO4, DOD |
 |
High-Resolution Neutron And X-Ray Joint Refined Structure Of High-Potential Iron-Sulfur Protein In The Oxidized State
Organism: Thermochromatium tepidum
Method: X-RAY DIFFRACTION, NEUTRON DIFFRACTION Resolution:0.66 Å, 1.20 Å Release Date: 2022-06-01 Classification: METAL BINDING PROTEIN Ligands: SF4, SO4, GOL, NH4, D3O, DOD |
 |
Crystallographic Structure Of Copper Amine Oxidase From Arthrobacter Glibiformis At Pd 7.4 Determined By Only Neutron Diffraction Data.
Organism: Arthrobacter globiformis
Method: NEUTRON DIFFRACTION Resolution:1.72 Å Release Date: 2022-04-20 Classification: OXIDOREDUCTASE Ligands: CU, DOD |
 |
Crystallographic Structure Of Copper Amine Oxidase From Arthrobacter Glibiformis At Pd 7.4 Determined By Both X-Ray And Neutron Diffraction Data At 1.72 Angstrom Resolution.
Organism: Arthrobacter globiformis
Method: X-RAY DIFFRACTION, NEUTRON DIFFRACTION Resolution:1.7200 Å, 1.7200 Å Release Date: 2022-04-20 Classification: OXIDOREDUCTASE Ligands: CU, NA, DOD |
 |
Organism: Gallus gallus
Method: X-RAY DIFFRACTION Resolution:1.42 Å Release Date: 2022-04-13 Classification: HYDROLASE Ligands: NI, CL, DOD |

