Search Count: 3,261
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Resolution:3.32 Å Release Date: 2026-01-14 Classification: DNA BINDING PROTEIN |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Resolution:3.56 Å Release Date: 2026-01-14 Classification: DNA BINDING PROTEIN |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Resolution:3.39 Å Release Date: 2026-01-14 Classification: DNA BINDING PROTEIN |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Resolution:2.94 Å Release Date: 2026-01-14 Classification: DNA BINDING PROTEIN |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Resolution:4.54 Å Release Date: 2026-01-14 Classification: DNA BINDING PROTEIN |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Resolution:4.37 Å Release Date: 2026-01-14 Classification: DNA BINDING PROTEIN |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Resolution:4.38 Å Release Date: 2026-01-14 Classification: DNA BINDING PROTEIN |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2026-01-14 Classification: DNA BINDING PROTEIN |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:3.00 Å Release Date: 2026-01-14 Classification: PROTEIN BINDING |
 |
Organism: Bacillus subtilis
Method: X-RAY DIFFRACTION Resolution:1.36 Å Release Date: 2026-01-07 Classification: DNA BINDING PROTEIN Ligands: ADP, MG, SO4 |
 |
Organism: Homo sapiens, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.64 Å Release Date: 2025-12-31 Classification: LIGASE Ligands: DMS, AMP |
 |
Dna Ligase 1 E346A/E592A In Complex With Nick Containing 3'-8Oxorg:A Captured At Post-Catalytic Stage
Organism: Homo sapiens, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:1.96 Å Release Date: 2025-12-31 Classification: LIGASE/DNA Ligands: AMP |
 |
Dna Ligase 1 E346A/E592A In Complex With Nick Containing 3'-8Oxorg:C Captured At Pre-Catalytic Stage
Organism: Homo sapiens, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.81 Å Release Date: 2025-12-31 Classification: LIGASE/DNA |
 |
Dna Ligase 1 E346A/E592A In Complex With Nick Containing 3'-8Oxodg:A Captured At Pre-Catalytic Stage
Organism: Homo sapiens, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.56 Å Release Date: 2025-12-31 Classification: LIGASE/DNA Ligands: DMS |
 |
Dna Ligase 1 E346A/E592A In Complex With Nick Containing 3'-8Oxodg:C Captured At Pre-Catalytic Stage
Organism: Homo sapiens, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.96 Å Release Date: 2025-12-31 Classification: LIGASE/DNA |
 |
Dna Ligase 1 Wild-Type In Complex With Nick Containing 3'-8Oxodg:C Captured At Pre-Catalytic Stage
Organism: Homo sapiens, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.76 Å Release Date: 2025-12-31 Classification: LIGASE/DNA |
 |
Organism: Acetivibrio thermocellus
Method: ELECTRON MICROSCOPY Release Date: 2025-12-24 Classification: DNA BINDING PROTEIN Ligands: ZN |
 |
Organism: Acetivibrio thermocellus
Method: ELECTRON MICROSCOPY Release Date: 2025-12-24 Classification: DNA BINDING PROTEIN Ligands: ZN, ANP |
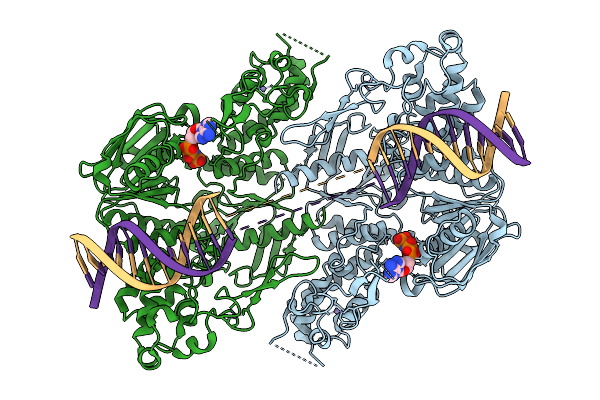 |
C. Thermocellum Uvra In Complex With Unmodified Dna And Amppnp (Basal Conformation)
Organism: Acetivibrio thermocellus, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2025-12-24 Classification: DNA BINDING PROTEIN Ligands: ZN, ANP |
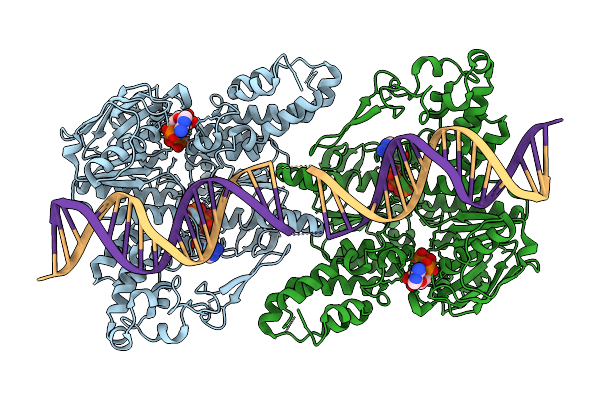 |
C. Thermocellum Uvra In Complex With Unmodified Dna And Amppnp (Atp-Bound Conformation 1)
Organism: Acetivibrio thermocellus, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2025-12-24 Classification: DNA BINDING PROTEIN Ligands: ZN, ANP |

