Search Count: 211
All
Selected
 |
Organism: Lomovskayavirus c31
Method: ELECTRON MICROSCOPY Release Date: 2026-02-11 Classification: DNA BINDING PROTEIN Ligands: ZN |
 |
Organism: Lomovskayavirus c31
Method: ELECTRON MICROSCOPY Release Date: 2026-02-11 Classification: DNA BINDING PROTEIN Ligands: ZN |
 |
Organism: Lomovskayavirus c31
Method: ELECTRON MICROSCOPY Release Date: 2026-02-11 Classification: DNA BINDING PROTEIN Ligands: ZN |
 |
Organism: Lomovskayavirus c31
Method: ELECTRON MICROSCOPY Release Date: 2026-02-11 Classification: DNA BINDING PROTEIN Ligands: ZN |
 |
Organism: Lomovskayavirus c31
Method: ELECTRON MICROSCOPY Release Date: 2026-02-11 Classification: DNA BINDING PROTEIN Ligands: ZN |
 |
Organism: Lomovskayavirus c31
Method: ELECTRON MICROSCOPY Release Date: 2026-02-11 Classification: RECOMBINATION Ligands: ZN |
 |
Organism: Lomovskayavirus c31
Method: ELECTRON MICROSCOPY Release Date: 2026-02-11 Classification: RECOMBINATION Ligands: ZN |
 |
Gcf1P, Multimerizes And Bridges The Mitochondrial Dna From Candida Albicans By A Specific Mechanism.
Organism: Candida albicans, Synthetic construct
Method: X-RAY DIFFRACTION, SOLUTION SCATTERING Resolution:2.90 Å Release Date: 2023-06-14 Classification: DNA BINDING PROTEIN Ligands: GOL |
 |
Structure Of Single Homo-Hexameric Holliday Junction Atp-Dependent Dna Helicase Ruvb Motor
Organism: Thermus thermophilus hb8
Method: ELECTRON MICROSCOPY Release Date: 2023-05-10 Classification: STRUCTURAL PROTEIN Ligands: AGS, MG, ADP |
 |
Organism: Thermus thermophilus hb8
Method: ELECTRON MICROSCOPY Release Date: 2023-05-10 Classification: STRUCTURAL PROTEIN Ligands: ADP, MG, AGS |
 |
Structure Of Rect Protein From Listeria Innoccua Phage A118 In Complex With 83-Mer Annealed Duplex
Organism: Listeria innocua clip11262, Escherichia virus m13
Method: ELECTRON MICROSCOPY Release Date: 2022-12-07 Classification: DNA BINDING PROTEIN/DNA |
 |
Structure Of Rect Protein From Listeria Innoccua Phage A118 In Complex With 83-Mer Ssdna
Organism: Listeria innocua clip11262
Method: ELECTRON MICROSCOPY Release Date: 2022-12-07 Classification: DNA BINDING PROTEIN |
 |
Ruvab Branch Migration Motor Complexed To The Holliday Junction - Ruvb Aaa+ State S1 [T2 Dataset]
Organism: Streptococcus thermophilus, Salmonella typhimurium, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2022-09-14 Classification: HYDROLASE Ligands: AGS, MG, ADP |
 |
Ruvab Branch Migration Motor Complexed To The Holliday Junction - Ruvb Aaa+ State S2 [T2 Dataset]
Organism: Streptococcus thermophilus, Salmonella typhimurium, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2022-09-14 Classification: HYDROLASE Ligands: AGS, MG, ADP |
 |
Ruvab Branch Migration Motor Complexed To The Holliday Junction - Ruvb Aaa+ State S3 [T2 Dataset]
Organism: Streptococcus thermophilus, Salmonella typhimurium, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2022-09-14 Classification: HYDROLASE Ligands: ADP, MG, AGS |
 |
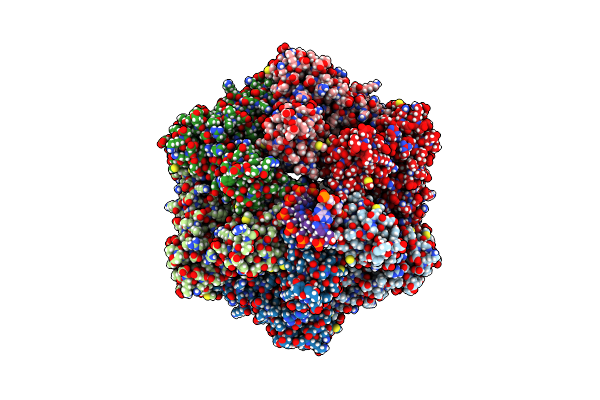
Ruvab Branch Migration Motor Complexed To The Holliday Junction - Ruvb Aaa+ State S4 [T2 Dataset]
Organism: Streptococcus thermophilus, Salmonella typhimurium, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2022-09-14 Classification: HYDROLASE Ligands: ADP, AGS, MG |
 |
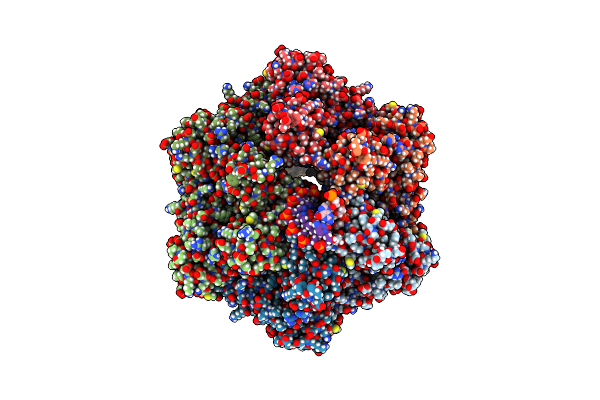
Ruvab Branch Migration Motor Complexed To The Holliday Junction - Ruvb Aaa+ State S5 [T2 Dataset]
Organism: Streptococcus thermophilus, Salmonella typhimurium, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2022-09-14 Classification: HYDROLASE Ligands: ADP, AGS, MG |
 |
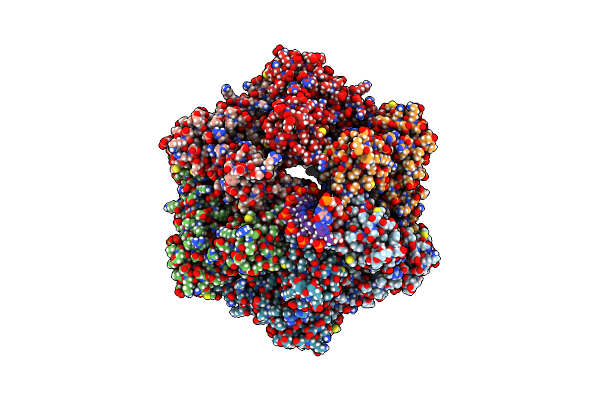
Ruvab Branch Migration Motor Complexed To The Holliday Junction - Ruvb Aaa+ State S0+A [T2 Dataset]
Organism: Streptococcus thermophilus, Salmonella typhimurium, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2022-09-14 Classification: HYDROLASE Ligands: AGS, MG, ADP |
 |
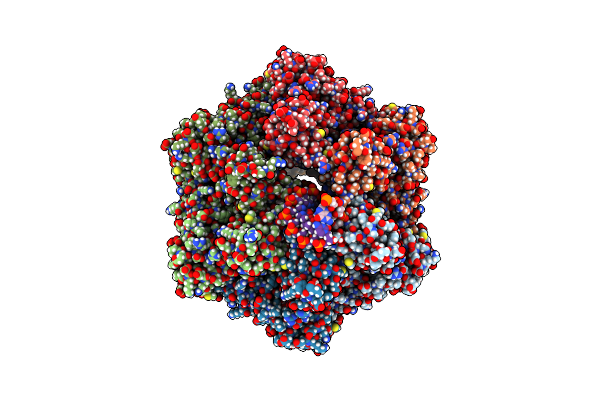
Ruvab Branch Migration Motor Complexed To The Holliday Junction - Ruvb Aaa+ State S0-A [T2 Dataset]
Organism: Streptococcus thermophilus, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2022-09-14 Classification: HYDROLASE Ligands: AGS, MG, ADP |
 |
Ruvab Branch Migration Motor Complexed To The Holliday Junction - Ruvb Aaa+ State S0+A [T1 Dataset]
Organism: Streptococcus thermophilus, Salmonella typhimurium, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2022-09-14 Classification: HYDROLASE Ligands: AGS, MG, ADP |

