Search Count: 1,05,966
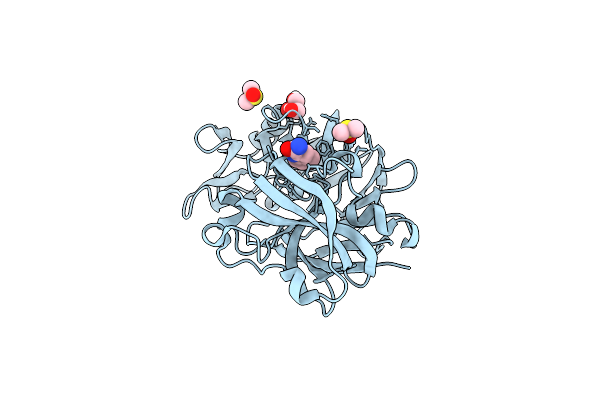 |
Crystal Structure Of Endothiapepsin In Complex With Fragment Eos102639 From Ecbl-96
Organism: Cryphonectria parasitica
Method: X-RAY DIFFRACTION Release Date: 2025-11-05 Classification: HYDROLASE Ligands: A1CE5, DMS, GOL |
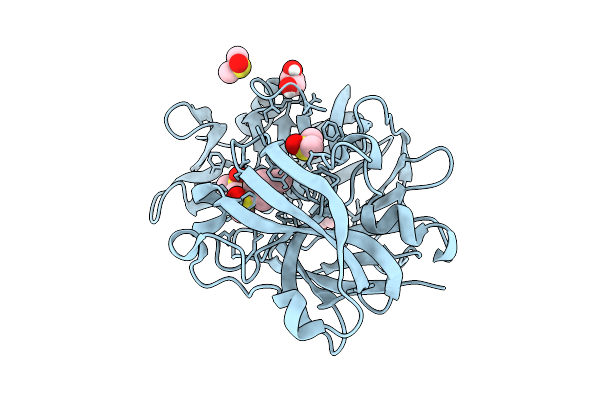 |
Crystal Structure Of Endothiapepsin In Complex With Fragment Eos102793 From Ecbl-96
Organism: Cryphonectria parasitica
Method: X-RAY DIFFRACTION Release Date: 2025-11-05 Classification: HYDROLASE Ligands: A1CFC, GOL, DMS |
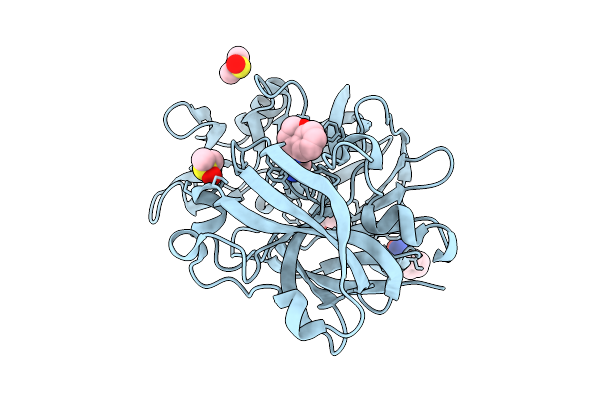 |
Crystal Structure Of Endothiapepsin In Complex With Fragment Eos102887 From Ecbl-96
Organism: Cryphonectria parasitica
Method: X-RAY DIFFRACTION Release Date: 2025-11-05 Classification: HYDROLASE Ligands: A1CFD, DMS, GOL |
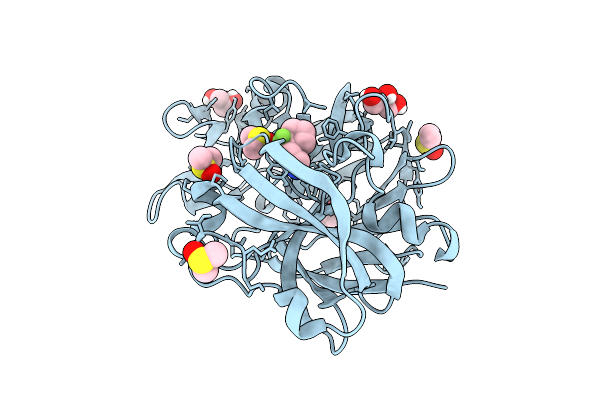 |
Crystal Structure Of Endothiapepsin In Complex With Fragment Eos103099 From Ecbl-96
Organism: Cryphonectria parasitica
Method: X-RAY DIFFRACTION Release Date: 2025-11-05 Classification: HYDROLASE Ligands: A1CDX, DMS, GOL |
 |
Crystal Structure Of Endothiapepsin In Complex With Fragment Eos103134 From Ecbl-96
Organism: Cryphonectria parasitica
Method: X-RAY DIFFRACTION Release Date: 2025-11-05 Classification: HYDROLASE Ligands: A1CDR, GOL |
 |
Crystal Structure Of Endothiapepsin In Complex With Fragment Eos102483 From Ecbl-96
Organism: Cryphonectria parasitica
Method: X-RAY DIFFRACTION Release Date: 2025-11-05 Classification: HYDROLASE Ligands: A1CFE |
 |
Crystal Structure Of Endothiapepsin In Complex With Fragment Eos102671 From Ecbl-96
Organism: Cryphonectria parasitica
Method: X-RAY DIFFRACTION Release Date: 2025-11-05 Classification: HYDROLASE Ligands: A1CFF |
 |
Crystal Structure Of Endothiapepsin In Complex With Fragment Eos102818 From Ecbl-96
Organism: Cryphonectria parasitica
Method: X-RAY DIFFRACTION Release Date: 2025-11-05 Classification: HYDROLASE Ligands: WQY |
 |
Crystal Structure Of Endothiapepsin In Complex With Fragment Eos102900 From Ecbl-96
Organism: Cryphonectria parasitica
Method: X-RAY DIFFRACTION Release Date: 2025-11-05 Classification: HYDROLASE Ligands: A1CFG |
 |
Crystal Structure Of Endothiapepsin In Complex With Fragment Eos103107 From Ecbl-96
Organism: Cryphonectria parasitica
Method: X-RAY DIFFRACTION Release Date: 2025-11-05 Classification: HYDROLASE Ligands: A1EHQ, DMS |
 |
Crystal Structure Of Endothiapepsin In Complex With Fragment Eos103253 From Ecbl-96
Organism: Cryphonectria parasitica
Method: X-RAY DIFFRACTION Release Date: 2025-11-05 Classification: HYDROLASE Ligands: A1CFM, DMS |
 |
Crystal Structure Of Endothiapepsin In Complex With Fragment Eos102489 From Ecbl-96
Organism: Cryphonectria parasitica
Method: X-RAY DIFFRACTION Release Date: 2025-11-05 Classification: HYDROLASE Ligands: A1CFR |
 |
Crystal Structure Of Endothiapepsin In Complex With Fragment Eos102680 From Ecbl-96
Organism: Cryphonectria parasitica
Method: X-RAY DIFFRACTION Release Date: 2025-11-05 Classification: HYDROLASE Ligands: A1CDO, DMS |
 |
Crystal Structure Of Endothiapepsin In Complex With Fragment Eos102843 From Ecbl-96
Organism: Cryphonectria parasitica
Method: X-RAY DIFFRACTION Release Date: 2025-11-05 Classification: HYDROLASE Ligands: A1CFS, DMS, ACT, GOL |
 |
Crystal Structure Of Endothiapepsin In Complex With Fragment Eos102933 From Ecbl-96
Organism: Cryphonectria parasitica
Method: X-RAY DIFFRACTION Release Date: 2025-11-05 Classification: HYDROLASE Ligands: WOY, DMS, GOL |
 |
Crystal Structure Of Endothiapepsin In Complex With Fragment Eos103110 From Ecbl-96
Organism: Cryphonectria parasitica
Method: X-RAY DIFFRACTION Release Date: 2025-11-05 Classification: HYDROLASE Ligands: A1CFT, DMS, PG5, GOL, ACT, PEG |
 |
Crystal Structure Of Endothiapepsin In Complex With Fragment Eos103290 From Ecbl-96
Organism: Cryphonectria parasitica
Method: X-RAY DIFFRACTION Release Date: 2025-11-05 Classification: HYDROLASE Ligands: A1CFV |
 |
Crystal Structure Of Endothiapepsin In Complex With Fragment Eos102546 From Ecbl-96
Organism: Cryphonectria parasitica
Method: X-RAY DIFFRACTION Release Date: 2025-11-05 Classification: HYDROLASE Ligands: A1CFW, DMS |
 |
Crystal Structure Of Endothiapepsin In Complex With Fragment Eos102688 From Ecbl-96
Organism: Cryphonectria parasitica
Method: X-RAY DIFFRACTION Release Date: 2025-11-05 Classification: HYDROLASE Ligands: DBP, GOL, DMS |
 |
Crystal Structure Of Endothiapepsin In Complex With Fragment Eos102857 From Ecbl-96
Organism: Cryphonectria parasitica
Method: X-RAY DIFFRACTION Release Date: 2025-11-05 Classification: HYDROLASE Ligands: A1CFX, GOL |

