Search Count: 415
All
Selected
 |
Crystal Structure Of Plasmoredoxin, A Disulfide Oxidoreductase From Plasmodium Falciparum Crystallized In The Presence Of Dithiothreitol (Dtt)
Organism: Plasmodium falciparum 3d7
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2025-11-12 Classification: OXIDOREDUCTASE |
 |
Crystal Structure Of Plasmoredoxin, A Disulfide Oxidoreductase From Plasmodium Falciparum
Organism: Plasmodium falciparum 3d7
Method: X-RAY DIFFRACTION Resolution:1.77 Å Release Date: 2024-11-06 Classification: OXIDOREDUCTASE |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2024-10-30 Classification: LIGASE Ligands: MG, NAD, FDA |
 |
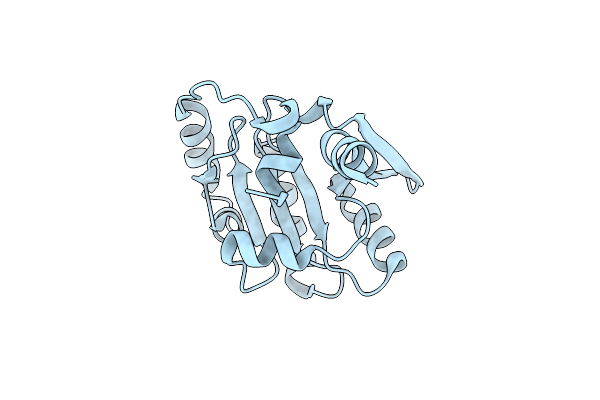
Crystal Structure Of Plasmoredoxin, A Disulfide Oxidoreductase From Plasmodium Falciparum Crystallized In The Presence Of Glutathione(Gsh)
Organism: Plasmodium falciparum 3d7
Method: X-RAY DIFFRACTION Resolution:1.40 Å Release Date: 2024-10-30 Classification: OXIDOREDUCTASE Ligands: SO4 |
 |
Crystal Structure Of Plasmoredoxin From Plasmodium Falciparum A Disulfide Oxidoreductase Protein Unique To Plasmodium Species
Organism: Plasmodium falciparum 3d7
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2024-10-16 Classification: OXIDOREDUCTASE |
 |
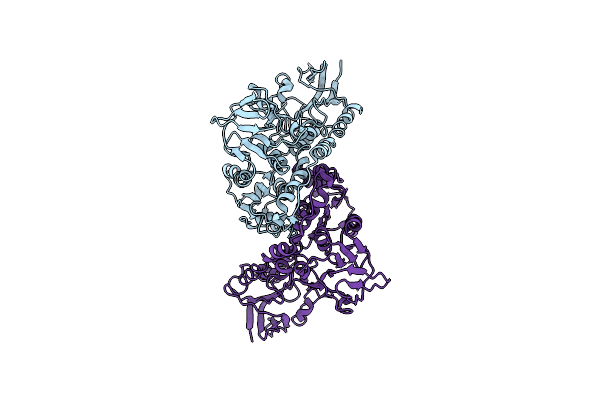
Organism: Escherichia coli k-12
Method: X-RAY DIFFRACTION Resolution:1.74 Å Release Date: 2024-09-11 Classification: OXIDOREDUCTASE/OXIDOREDUCTASE INHIBITOR Ligands: 5V9, CU |
 |
Crystal Structure Of Mycothiol Disulfide Reductase Mtr From Mycobacterium Smegmatis
Organism: Mycolicibacterium smegmatis mc2 155
Method: X-RAY DIFFRACTION Resolution:4.70 Å Release Date: 2024-03-13 Classification: OXIDOREDUCTASE |
 |
E.Coli Dsba In Complex With N-(2-Fluorophenyl)-5-Methylisoxazole-3-Carboxamide
Organism: Escherichia coli k-12
Method: X-RAY DIFFRACTION Resolution:1.57 Å Release Date: 2023-06-21 Classification: OXIDOREDUCTASE/INHIBITOR Ligands: SW0, EDO, NA, CU |
 |
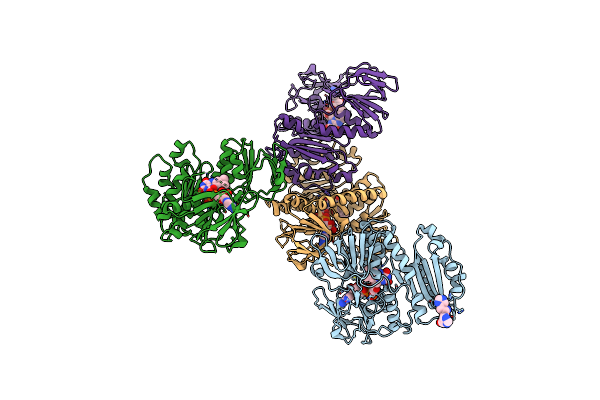
E.Coli Dsba In Complex With 4-Phenyl-2-(3-Phenylpropyl)Thiazole-5-Carboxylic Acid
Organism: Escherichia coli k-12
Method: X-RAY DIFFRACTION Resolution:1.99 Å Release Date: 2023-04-12 Classification: OXIDOREDUCTASE/OXIDOREDUCTASE INHIBITOR Ligands: QVP, CU |
 |
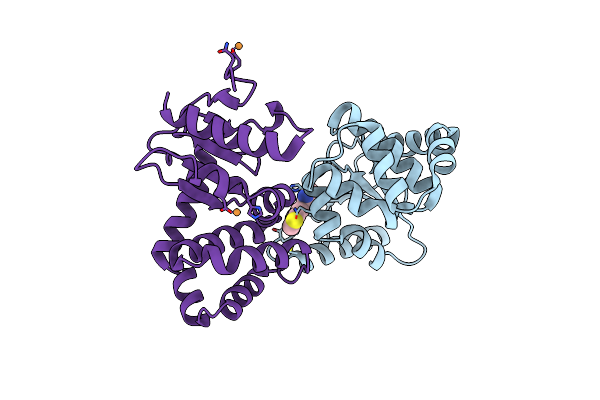
Crystal Structure Of The Disulfide Reductase Mera From Staphylococcus Aureus
Organism: Staphylococcus aureus
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2023-03-01 Classification: OXIDOREDUCTASE Ligands: FAD, HIS |
 |
Crystal Structure Of E.Coli Dsba In Complex With Compound Mips-0001897 (Compound 1)
Organism: Escherichia coli (strain k12)
Method: X-RAY DIFFRACTION Resolution:1.95 Å Release Date: 2023-02-08 Classification: OXIDOREDUCTASE/OXIDOREDUCTASE inhibitor Ligands: 5VA, CU |
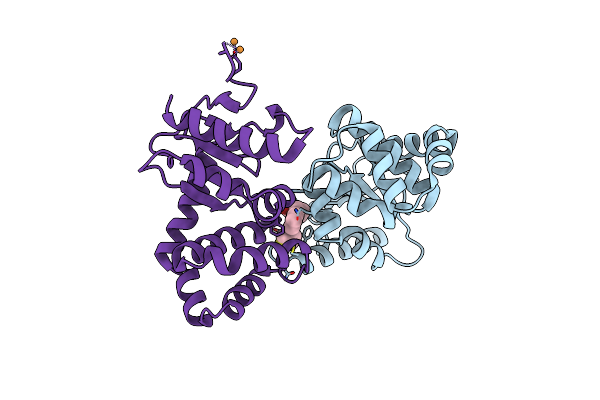 |
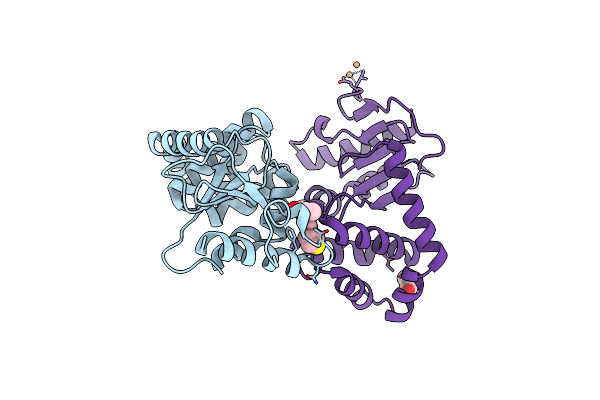
Crystal Structure Of E.Coli Dsba In Complex With Compound Mips-0001877 (Compound 39)
Organism: Escherichia coli (strain k12)
Method: X-RAY DIFFRACTION Resolution:1.59 Å Release Date: 2023-02-08 Classification: OXIDOREDUCTASE/OXIDOREDUCTASE inhibitor Ligands: 648, CU |
 |
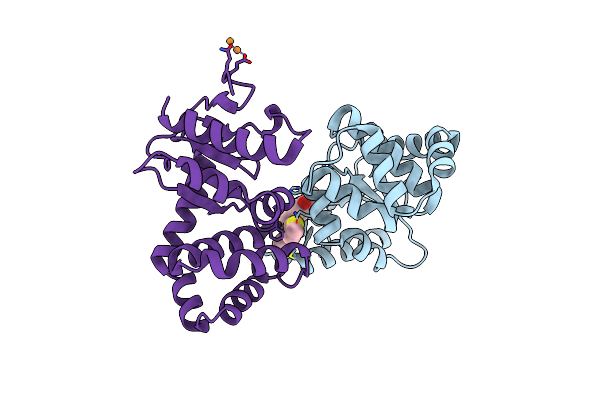
Crystal Structure Of E.Coli Dsba In Complex With Compound Mips-0001886 (Compound 38)
Organism: Escherichia coli (strain k12)
Method: X-RAY DIFFRACTION Resolution:1.76 Å Release Date: 2023-02-08 Classification: OXIDOREDUCTASE/OXIDOREDUCTASE inhibitor Ligands: 5VB, CU, GOL |
 |
Crystal Structure Of E.Coli Dsba In Complex With Compound Mips-0001896 (Compound 72)
Organism: Escherichia coli (strain k12)
Method: X-RAY DIFFRACTION Resolution:1.62 Å Release Date: 2023-02-08 Classification: OXIDOREDUCTASE/OXIDOREDUCTASE inhibitor Ligands: 62J, CU |
 |
Organism: Escherichia coli k-12
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2022-12-07 Classification: OXIDOREDUCTASE Ligands: CU, 010 |
 |
Organism: Escherichia coli k-12
Method: X-RAY DIFFRACTION Resolution:1.47 Å Release Date: 2022-12-07 Classification: OXIDOREDUCTASE Ligands: IMD |
 |
Organism: Escherichia coli k-12
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2022-12-07 Classification: OXIDOREDUCTASE Ligands: BYZ, PGE |
 |
Organism: Escherichia coli k-12
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2022-12-07 Classification: OXIDOREDUCTASE Ligands: PKN, CU |
 |
Crystal Structure Of Ecdsba In A Complex With (1-Methyl-1H-Pyrazol-5-Yl)Methanamine
Organism: Escherichia coli k-12
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2022-12-07 Classification: OXIDOREDUCTASE Ligands: Q2I, CU |
 |
Organism: Escherichia coli k-12
Method: X-RAY DIFFRACTION Resolution:1.85 Å Release Date: 2022-12-07 Classification: OXIDOREDUCTASE Ligands: Q2O, CU |

