Search Count: 22,746
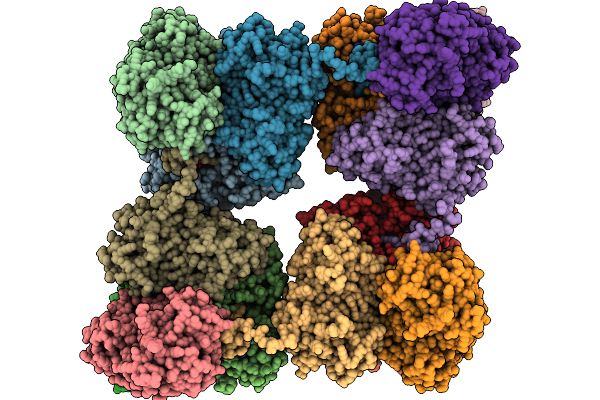 |
Photosynthetic A8B8 Glyceraldehyde-3-Phosphate Dehydrogenase (Minor Conformer) From Spinacia Oleracea.
Organism: Spinacia oleracea
Method: ELECTRON MICROSCOPY Release Date: 2026-01-21 Classification: OXIDOREDUCTASE Ligands: NAD |
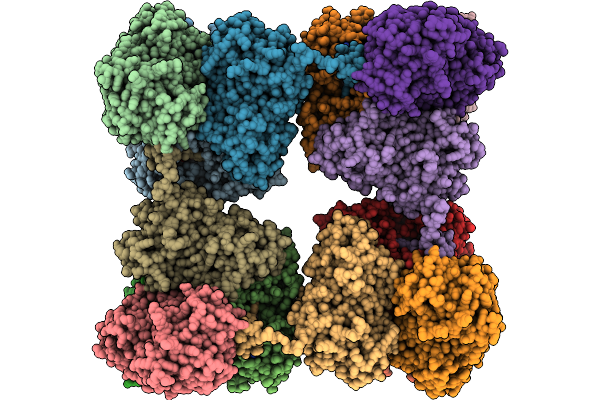 |
Photosynthetic A8B8 Glycerldeyde-3-Phosphate Dehydrogenase Hexadecamer (Major Conformer) From Spinacia Oleracea.
Organism: Spinacia oleracea
Method: ELECTRON MICROSCOPY Release Date: 2026-01-21 Classification: OXIDOREDUCTASE Ligands: NAD |
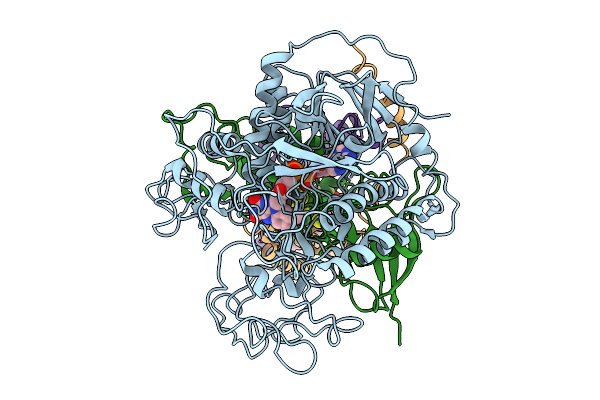 |
Cryo-Em Structure Of Saccharomyces Cerevisiae Mitochondrial Respiratory Complex Ii In Cyclobutrifluram-Bound State
Organism: Saccharomyces cerevisiae s288c
Method: ELECTRON MICROSCOPY Release Date: 2026-01-21 Classification: MEMBRANE PROTEIN Ligands: FAD, A1EKU, FES, SF4, F3S, PEE |
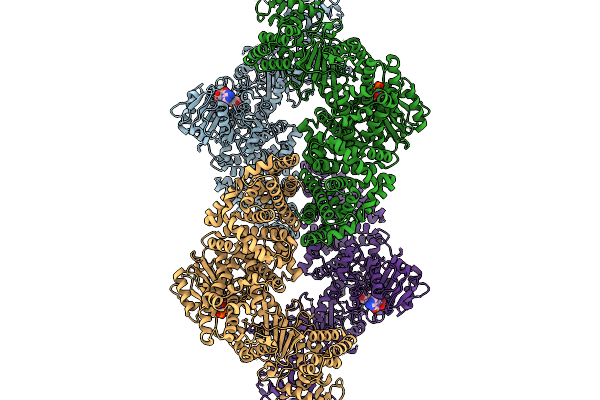 |
Cryoem Map Of The Large Glutamate Dehydrogenase Composed Of 180 Kda Subunits From Mycobacterium Smegmatis Obtained In The Presence Of Nad+ And L-Glutamate. Open Tetramer.
Organism: Mycolicibacterium smegmatis
Method: ELECTRON MICROSCOPY Release Date: 2026-01-14 Classification: OXIDOREDUCTASE Ligands: NAD |
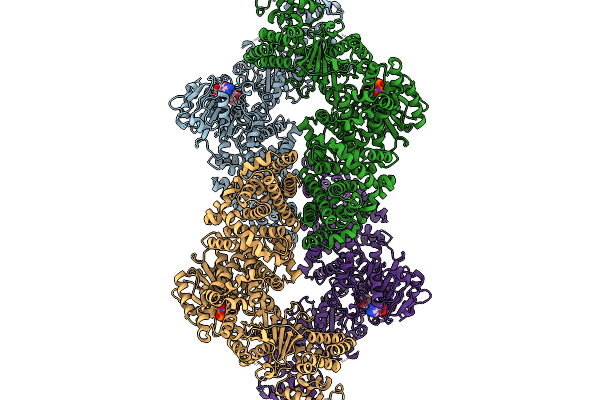 |
Cryoem Map Of The Large Glutamate Dehydrogenase Composed Of 180 Kda Subunits From Mycobacterium Smegmatis Obtained In The Presence Of Nad+ And L-Glutamate. Closed1 Tetramer.
Organism: Mycolicibacterium smegmatis
Method: ELECTRON MICROSCOPY Release Date: 2026-01-14 Classification: OXIDOREDUCTASE Ligands: NAD |
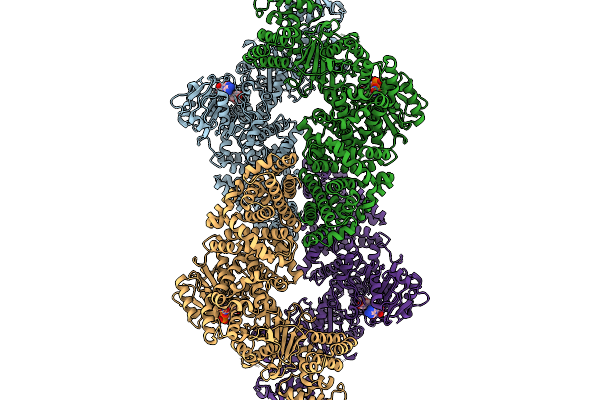 |
Cryoem Map Of The Large Glutamate Dehydrogenase Composed Of 180 Kda Subunits From Mycobacterium Smegmatis Obtained In The Presence Of Nad+ And L-Glutamate. Closed2 Tetramer
Organism: Mycolicibacterium smegmatis
Method: ELECTRON MICROSCOPY Release Date: 2026-01-14 Classification: OXIDOREDUCTASE Ligands: NAD |
 |
Cryoem Map Of The Large Glutamate Dehydrogenase Composed Of 180 Kda Subunits From Mycobacterium Smegmatis Obtained In The Presence Of Nad+ And L-Glutamate. Empty Monomer.
Organism: Mycolicibacterium smegmatis
Method: ELECTRON MICROSCOPY Release Date: 2026-01-14 Classification: OXIDOREDUCTASE |
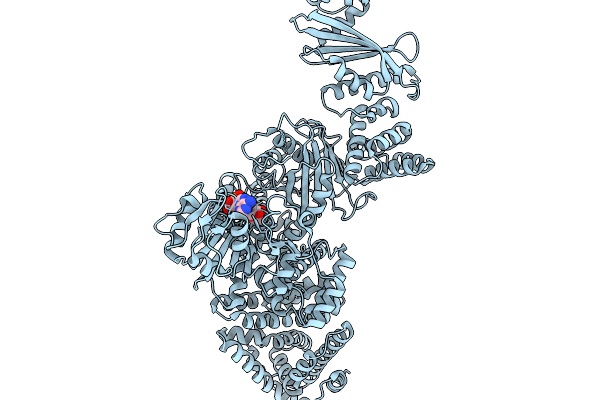 |
Cryoem Map Of The Large Glutamate Dehydrogenase Composed Of 180 Kda Subunits From Mycobacterium Smegmatis Obtained In The Presence Of Nad+ And L-Glutamate. Cofactor-Monomer.
Organism: Mycolicibacterium smegmatis
Method: ELECTRON MICROSCOPY Release Date: 2026-01-14 Classification: OXIDOREDUCTASE Ligands: NAD |
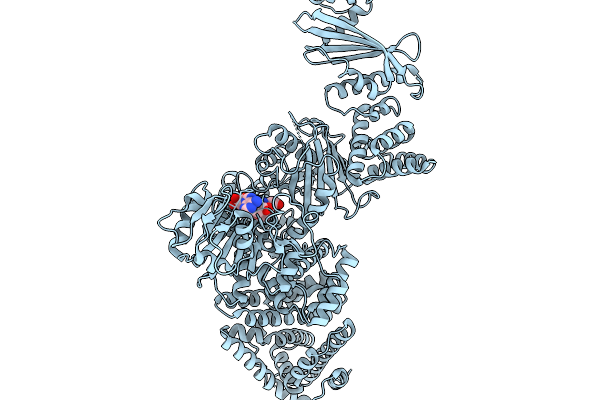 |
Cryoem Map Of The Large Glutamate Dehydrogenase Composed Of 180 Kda Subunits From Mycobacterium Smegmatis Obtained In The Presence Of Nad+ And L-Glutamate. Cofactor/Ligand-Monomer
Organism: Mycolicibacterium smegmatis
Method: ELECTRON MICROSCOPY Release Date: 2026-01-14 Classification: OXIDOREDUCTASE Ligands: GLU, NAD |
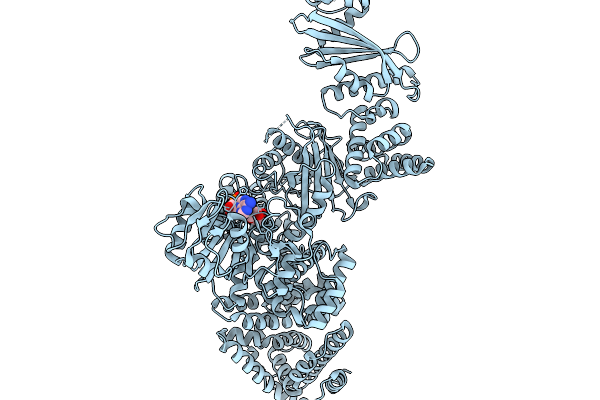 |
Cryoem Map Of The Large Glutamate Dehydrogenase Composed Of 180 Kda Subunits From Mycobacterium Smegmatis Obtained In The Presence Of Nad+ And L-Glutamate. Total-Monomer
Organism: Mycolicibacterium smegmatis
Method: ELECTRON MICROSCOPY Release Date: 2026-01-14 Classification: OXIDOREDUCTASE Ligands: NAD |
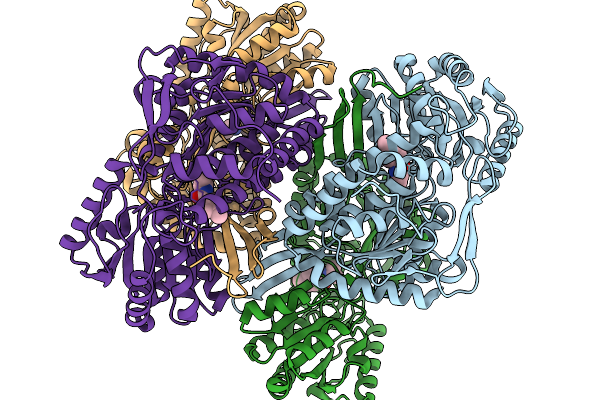 |
Co-Crystal Structure Of Phbdd-L163G With Product N-Isopropyl-4-Methylbenzamide
Organism: Pseudomonas putida
Method: X-RAY DIFFRACTION Resolution:1.83 Å Release Date: 2026-01-14 Classification: OXIDOREDUCTASE Ligands: A1EBJ |
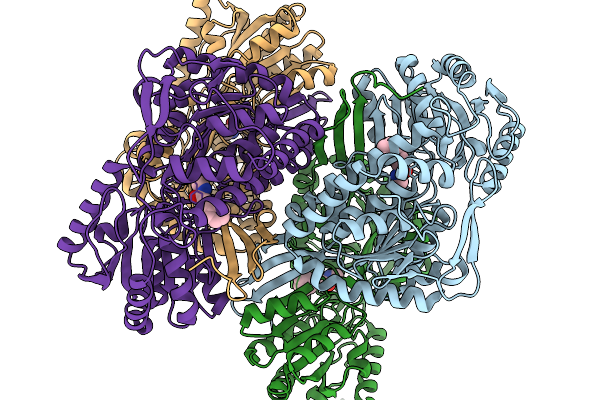 |
Organism: Pseudomonas putida
Method: X-RAY DIFFRACTION Resolution:2.06 Å Release Date: 2026-01-14 Classification: OXIDOREDUCTASE Ligands: A1EBQ |
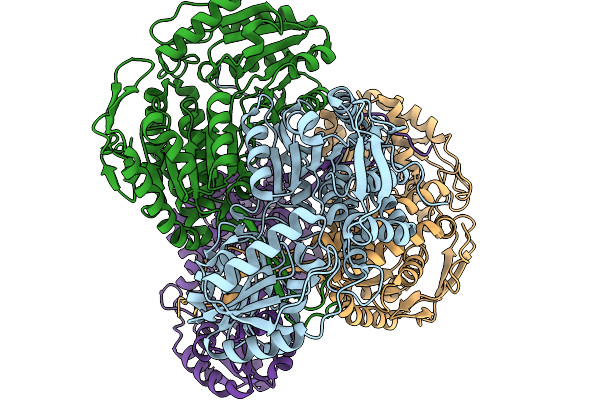 |
Organism: Pseudomonas putida
Method: X-RAY DIFFRACTION Resolution:1.89 Å Release Date: 2026-01-14 Classification: OXIDOREDUCTASE |
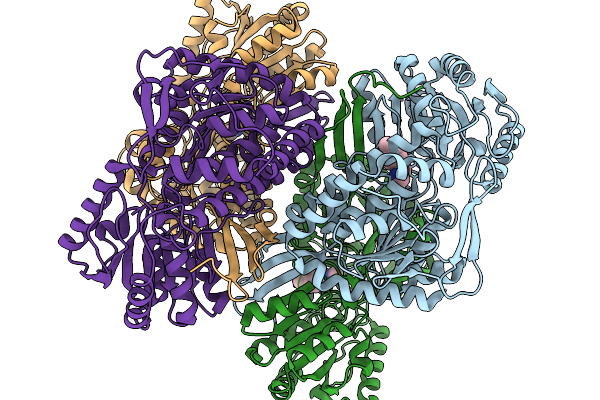 |
Organism: Pseudomonas putida
Method: X-RAY DIFFRACTION Resolution:2.21 Å Release Date: 2026-01-14 Classification: OXIDOREDUCTASE Ligands: A1EC9 |
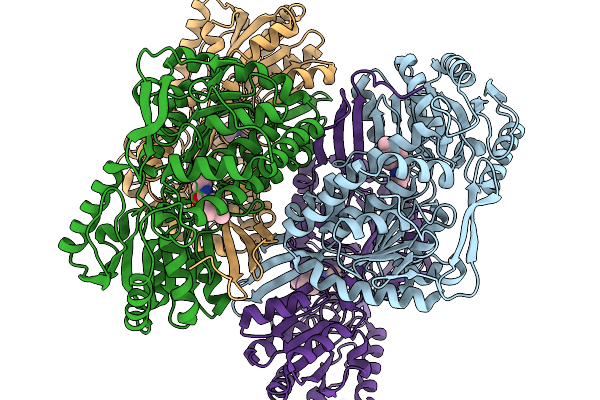 |
Co-Crystal Structure Of Phbdd-M1-L163G-T234A With N-Cyclopropyl-4-Methylbenzamide
Organism: Pseudomonas putida
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2026-01-14 Classification: OXIDOREDUCTASE Ligands: A1EDB |
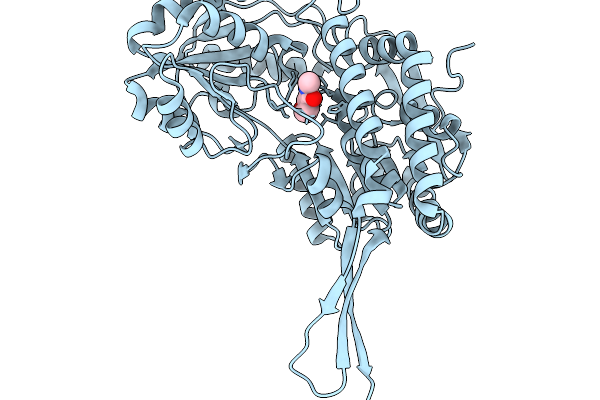 |
Organism: Pseudomonas putida
Method: X-RAY DIFFRACTION Resolution:1.93 Å Release Date: 2026-01-14 Classification: OXIDOREDUCTASE Ligands: A1EDA, SO4 |
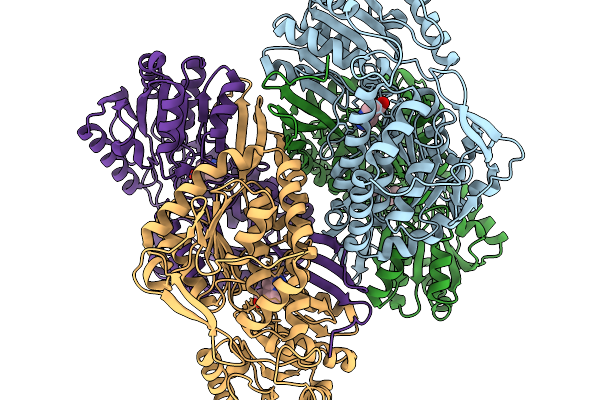 |
Co-Crystal Structure Of Phbdd-M1 With 1H-Pyrrolo[2,3-B]Pyridine-4-Carbaldehyde
Organism: Pseudomonas putida
Method: X-RAY DIFFRACTION Resolution:2.14 Å Release Date: 2026-01-14 Classification: OXIDOREDUCTASE Ligands: A1EDC |
 |
Organism: Pseudomonas putida
Method: X-RAY DIFFRACTION Resolution:3.12 Å Release Date: 2026-01-14 Classification: OXIDOREDUCTASE |
 |
Organism: Staphylococcus aureus
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2026-01-14 Classification: OXIDOREDUCTASE Ligands: ACY, CU1, NA, PGE |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2026-01-14 Classification: MEMBRANE PROTEIN Ligands: SF4, FMN, PEE, 8Q1, NDP, FES, PLX, ZN, CDL, DGT, MG |

