Search Count: 5,247
All
Selected
 |
Fe/Alpha-Kg-Dependent Decarboxylase Trah In Complexes With Mn2+, Alpha-Kg, And Substrate
Organism: Penicillium crustosum
Method: X-RAY DIFFRACTION Resolution:2.89 Å Release Date: 2026-03-04 Classification: OXIDOREDUCTASE Ligands: AKG, MN, A1I30, PEG |
 |
Crystal Structure Of Fe/Alphakg-Dependent Decarboxylase Trah In Complexes With Mn2+
Organism: Penicillium crustosum
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2026-03-04 Classification: OXIDOREDUCTASE Ligands: MN |
 |
Fe/Alpha-Kg-Dependent Decarboxylase Trah In Complexes With Mn2+, Alpha-Kg, And Substrate
Organism: Penicillium crustosum
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2026-03-04 Classification: OXIDOREDUCTASE Ligands: MN, AKG, A1I31, 1PG, PGE |
 |
Crystal Structure Of A Polyketide Decarboxylase Abx(+)O From Actinomycetes Sp. Ma7150
Organism: Actinomycetota bacterium
Method: X-RAY DIFFRACTION Resolution:1.99 Å Release Date: 2026-03-04 Classification: BIOSYNTHETIC PROTEIN Ligands: GOL, PO4 |
 |
Organism: Salmonella enterica subsp. enterica serovar typhimurium
Method: X-RAY DIFFRACTION Resolution:1.67 Å Release Date: 2026-02-11 Classification: LYASE Ligands: PUT, EDO, DMS, ACT, AG2, CL, NA |
 |
Crystal Structure Of None-Heme Iron Enzyme (Tqam) From Trichoderma Atroviride Bound With Iron And 2-Aminoisobutyric Acid
Organism: Trichoderma atroviride
Method: X-RAY DIFFRACTION Resolution:2.15 Å Release Date: 2026-02-04 Classification: METAL BINDING PROTEIN Ligands: FE, AIB |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2026-02-04 Classification: CYTOSOLIC PROTEIN Ligands: NAP |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2026-02-04 Classification: CYTOSOLIC PROTEIN Ligands: NAP |
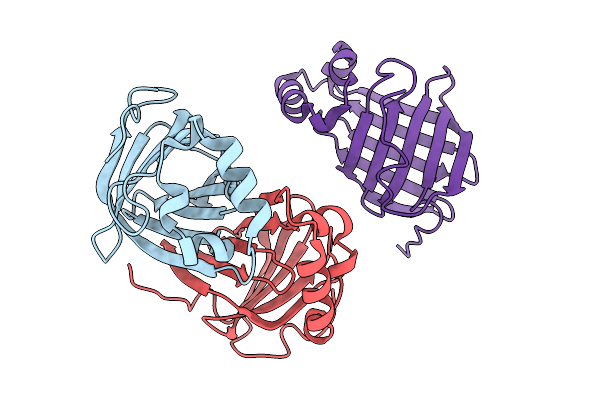 |
Crystal Structure Of Phenolic Acid Decarboxylase Mav Pad (Mycobacterium Avium)
Organism: Mycobacterium avium
Method: X-RAY DIFFRACTION Resolution:1.91 Å Release Date: 2026-01-28 Classification: OXIDOREDUCTASE |
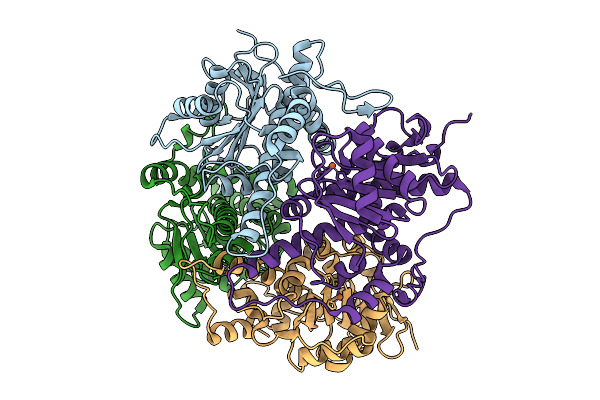 |
Crystal Structure Of None-Heme Iron Enzyme (Tqam) From Trichoderma Atroviride Bound With Iron
Organism: Trichoderma atroviride
Method: X-RAY DIFFRACTION Resolution:2.60 Å Release Date: 2026-01-28 Classification: METAL BINDING PROTEIN Ligands: FE |
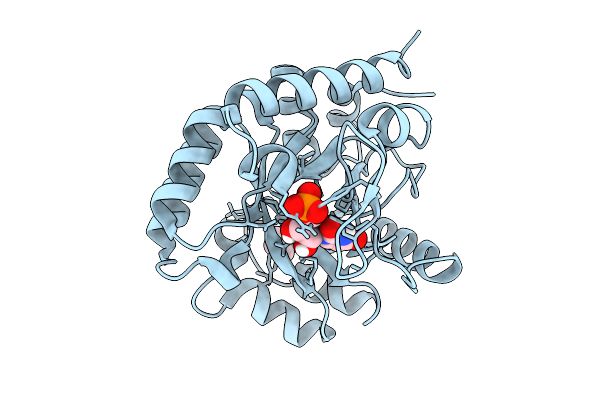 |
Variant T321A Of Orotidine 5'-Monophosphate Decarboxylase-Domain Of Human Umps In Complex With The Product Ump At 0.95 Angstrom Resolution
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:0.95 Å Release Date: 2026-01-21 Classification: LYASE Ligands: U5P, PRO |
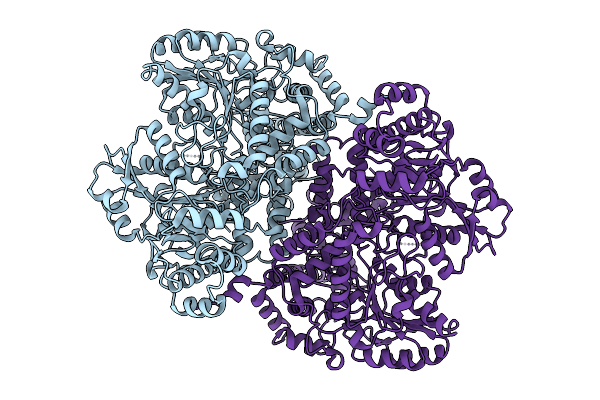 |
Organism: Escherichia coli str. k-12 substr. dh10b
Method: ELECTRON MICROSCOPY Resolution:2.32 Å Release Date: 2025-12-31 Classification: OXIDOREDUCTASE |
 |
Organism: Lacticaseibacillus rhamnosus
Method: X-RAY DIFFRACTION Resolution:2.09 Å Release Date: 2025-12-24 Classification: LYASE |
 |
Organism: Lacticaseibacillus rhamnosus
Method: X-RAY DIFFRACTION Resolution:2.64 Å Release Date: 2025-12-24 Classification: LYASE |
 |
Organism: Lacticaseibacillus rhamnosus
Method: X-RAY DIFFRACTION Resolution:2.69 Å Release Date: 2025-12-24 Classification: LYASE Ligands: PLP |
 |
Organism: Lacticaseibacillus rhamnosus
Method: X-RAY DIFFRACTION Resolution:1.74 Å Release Date: 2025-12-24 Classification: LYASE |
 |
Crystal Structure Of A Polyketide Abm/Scha-Like Domain-Containing Protein Whie-Orfi From Streptomyces Coelicolor
Organism: Streptomyces coelicolor
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2025-11-26 Classification: ANTIBIOTIC |
 |
Cryo Em Structure Of The Closed Tetramer Of Rv2531C From Mycobacterium Tuberculosis
Organism: Mycobacterium tuberculosis h37rv
Method: ELECTRON MICROSCOPY Resolution:3.30 Å Release Date: 2025-11-26 Classification: LYASE |
 |
Organism: Mycobacterium tuberculosis h37rv
Method: ELECTRON MICROSCOPY Resolution:2.80 Å Release Date: 2025-11-26 Classification: LYASE Ligands: PLP |
 |
Cryo Em Structure Of The Open Tetramer Of Rv2531C From Mycobacterium Tuberculosis.
Organism: Mycobacterium tuberculosis h37rv
Method: ELECTRON MICROSCOPY Resolution:3.40 Å Release Date: 2025-11-26 Classification: LYASE |

