Search Count: 76
 |
Organism: Saccharomyces cerevisiae
Method: ELECTRON MICROSCOPY Release Date: 2025-12-17 Classification: MEMBRANE PROTEIN Ligands: MG, DD9, D10, HP6, XKP, PEF, PLM, DCR |
 |
Structure Of The Double Cys-Substituted Cross-Linked Acrb Variant S562C_T837C
Organism: Escherichia coli k-12, Synthetic construct
Method: X-RAY DIFFRACTION Release Date: 2025-11-19 Classification: TRANSPORT PROTEIN Ligands: LMT, D10, GOL, D12, HEX, C14, OCT, DD9, SO4 |
 |
Cryo-Em Structure Of The Wild-Type Human Serotonin Transporter Complexed With S-Ketamine
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-11-12 Classification: TRANSPORT PROTEIN Ligands: CL, NA, Y01, LMT, NAG, JC9, DD9, D12, D10, OCT |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-07-16 Classification: MEMBRANE PROTEIN Ligands: TAU, D12, OCT, C14, DD9, HP6, D10, HEX, CL, NA, NAG |
 |
Crystal Structure Of Bax Core Domain Bh3-Groove Dimer - Hexameric Fraction With 2-Stearoyl Lysopc
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2023-12-27 Classification: APOPTOSIS Ligands: 1GP, EDO, OCT, D12, DD9 |
 |
Cryo-Em Structure Of Human Glycine Receptor Alpha1-Beta Heteromer, Glycine-Bound State 2(Expanded Open)
Organism: Homo sapiens, Aequorea victoria
Method: ELECTRON MICROSCOPY Release Date: 2023-10-11 Classification: MEMBRANE PROTEIN Ligands: NAG, HEX, HP6, UND, OCT, GLY, DD9, NBU, D10 |
 |
Cryo-Em Structure Of Human Glycine Receptor Alpha1-Beta Heteromer, Apo State
Organism: Homo sapiens, Aequorea victoria
Method: ELECTRON MICROSCOPY Release Date: 2023-10-11 Classification: MEMBRANE PROTEIN Ligands: NAG, HEX, UND, DD9, NBU, HP6, D10, CL |
 |
Cryo-Em Structure Of Human Glycine Receptor Alpha-1 Beta Heteromer, Glycine-Bound State3(Desensitized State)
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2023-10-11 Classification: MEMBRANE PROTEIN Ligands: NAG, LNK, HEX, NBU, OCT, DD9 |
 |
Cryo-Em Structure Of Human Glycine Receptor Alpha1-Beta Heteromer, Glycine-Bound State1(Open State)
Organism: Homo sapiens, Aequorea victoria
Method: ELECTRON MICROSCOPY Release Date: 2023-10-11 Classification: MEMBRANE PROTEIN Ligands: GLY, NAG, DD9, HP6, HEX, UND, NBU, D10, OCT |
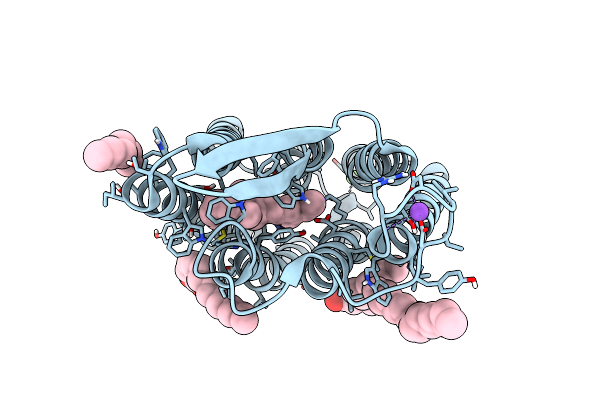 |
Room Temperature Structure Of Archaerhodopsin-3 Obtained 110 Ns After Photoexcitation
Organism: Halorubrum sodomense
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2023-06-14 Classification: PROTON TRANSPORT Ligands: RET, DD9, R16, PLM, CA, NA, MG, CL |
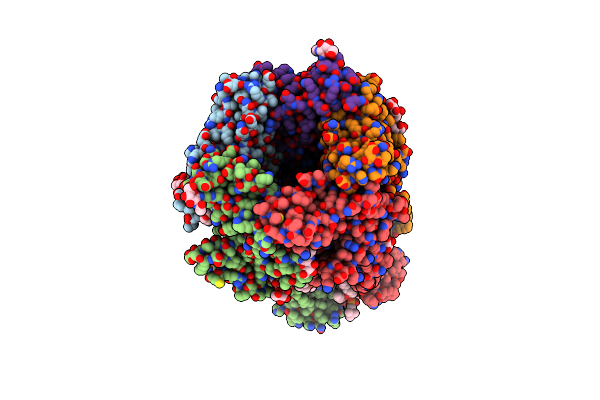 |
Cryo-Em Structure Of Torpedo Nicotinic Acetylcholine Receptor In Complex With Rocuronium, Pore-Blocked State
Organism: Tetronarce californica
Method: ELECTRON MICROSCOPY Release Date: 2023-06-07 Classification: TRANSPORT PROTEIN/INHIBITOR Ligands: RBR, POV, CLR, NAG, DD9 |
 |
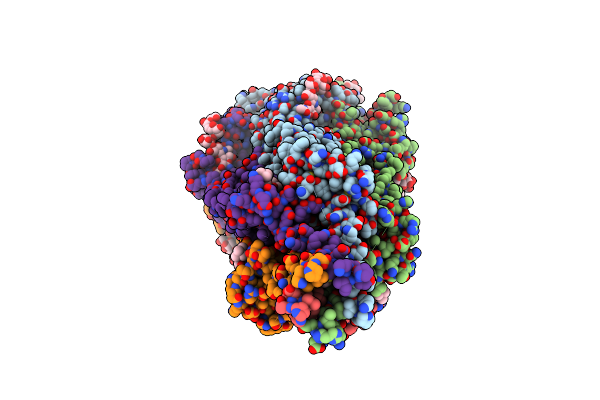
Cryo-Em Structure Of Torpedo Nicotinic Acetylcholine Receptor In Complex With Etomidate, Desensitized-Like State
Organism: Tetronarce californica
Method: ELECTRON MICROSCOPY Release Date: 2023-06-07 Classification: TRANSPORT PROTEIN/INHIBITOR Ligands: V8D, CHT, CLR, NAG, POV, DD9 |
 |
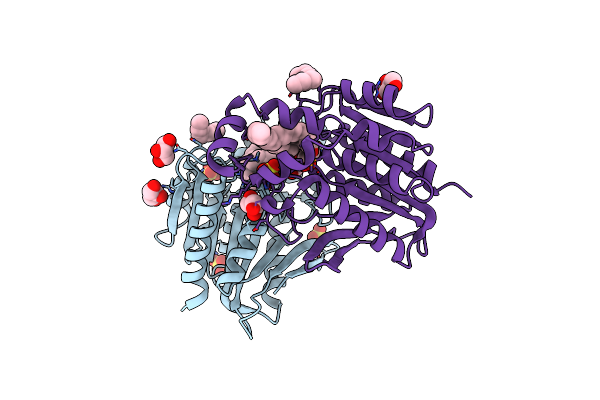
Cryo-Em Structure Of Torpedo Nicotinic Acetylcholine Receptor In Complex With Succinylcholine, Desensitized-Like State
Organism: Tetronarce californica
Method: ELECTRON MICROSCOPY Release Date: 2023-06-07 Classification: TRANSPORT PROTEIN/INHIBITOR Ligands: SCK, CLR, NAG, POV, DD9 |
 |
A Thermostable Lipase From Thermoanaerobacter Thermohydrosulfuricus In Complex A Monoacylglycerol Intermediate
Organism: Thermoanaerobacter thermohydrosulfuricus
Method: X-RAY DIFFRACTION Resolution:1.91 Å Release Date: 2023-05-10 Classification: HYDROLASE Ligands: STE, OCT, GOL, SO4, CL, DD9, D10 |
 |
Organism: Saccharomyces cerevisiae
Method: ELECTRON MICROSCOPY Release Date: 2023-03-29 Classification: TRANSFERASE Ligands: DD9, D10, C14, HP6, D12, XKP |
 |
Organism: Saccharomyces cerevisiae
Method: ELECTRON MICROSCOPY Release Date: 2023-03-29 Classification: TRANSFERASE Ligands: DD9, D10, C14, HP6, D12, XKP |
 |
Organism: Severe acute respiratory syndrome coronavirus 2
Method: X-RAY DIFFRACTION Resolution:2.95 Å Release Date: 2022-10-19 Classification: VIRAL PROTEIN Ligands: DD9, OCT |
 |
Organism: Tetronarce californica
Method: ELECTRON MICROSCOPY Resolution:2.50 Å Release Date: 2022-03-09 Classification: TRANSPORT PROTEIN Ligands: POV, CLR, NAG, DD9 |
 |
Cryo-Em Structure Of Torpedo Acetylcholine Receptor In Apo Form With Added Cholesterol
Organism: Tetronarce californica
Method: ELECTRON MICROSCOPY Resolution:2.74 Å Release Date: 2022-03-09 Classification: TRANSPORT PROTEIN Ligands: POV, CLR, NAG, DD9 |
 |
Cryo-Em Structure Of Torpedo Acetylcholine Receptor In Complex With Carbachol, Desensitized State
Organism: Tetronarce californica
Method: ELECTRON MICROSCOPY Resolution:2.77 Å Release Date: 2022-03-09 Classification: TRANSPORT PROTEIN Ligands: CCE, CLR, POV, NAG, DD9 |

