Search Count: 16
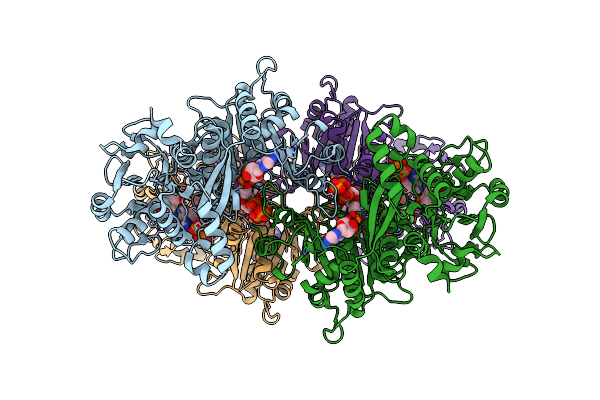 |
Organism: Drosophila melanogaster
Method: ELECTRON MICROSCOPY Release Date: 2024-11-27 Classification: LIGASE Ligands: DON, MG, DAT, DGT, 5ZL |
 |
Organism: Drosophila melanogaster
Method: ELECTRON MICROSCOPY Release Date: 2024-11-27 Classification: LIGASE Ligands: DON, MG, DAT, DGT, 5ZL |
 |
Organism: Giardia intestinalis
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2024-10-30 Classification: TRANSFERASE Ligands: DAT, D5M |
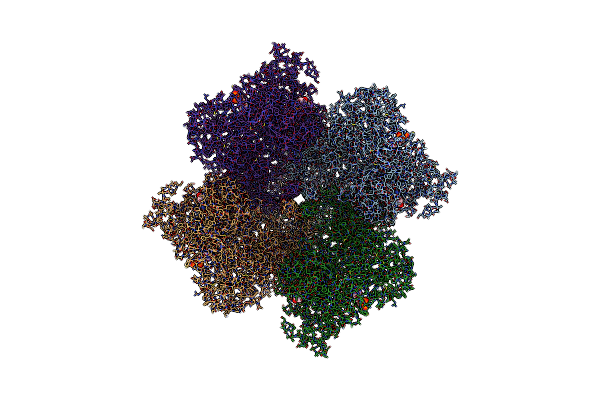 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2024-08-14 Classification: MEMBRANE PROTEIN Ligands: APR, DAT, ZN, CA |
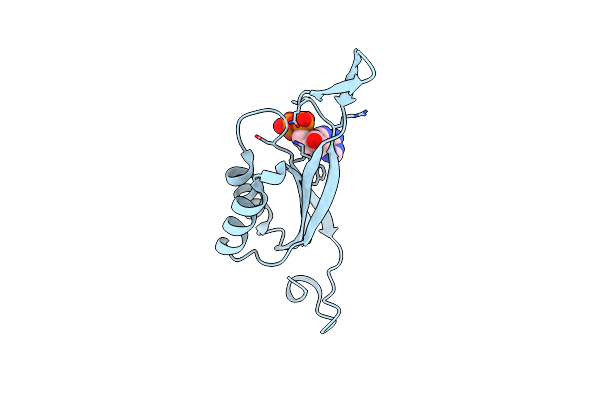 |
Glnk1 From Methanothermococcus Thermolithotrophicus With Dadp At A Resolution Of 1.94 A
Organism: Methanothermococcus thermolithotrophicus dsm 2095
Method: X-RAY DIFFRACTION Resolution:1.94 Å Release Date: 2021-10-06 Classification: SIGNALING PROTEIN Ligands: DAT, CL |
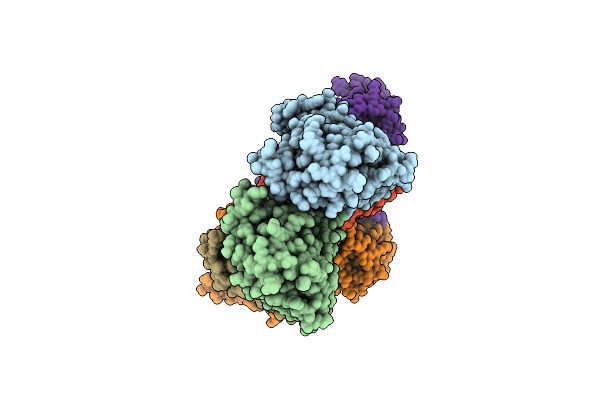 |
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:3.00 Å Release Date: 2016-12-07 Classification: HYDROLASE Ligands: DAT |
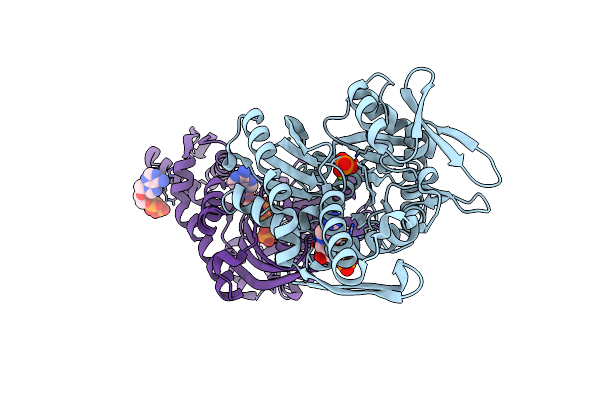 |
Crystal Structure Of Human Grp78 (70Kda Heat Shock Protein 5 / Bip) Atpase Domain In Complex With 2'-Deoxy-Adp And Inorganic Phosphate
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2016-06-15 Classification: CHAPERONE Ligands: MG, PO4, DTP, DAT |
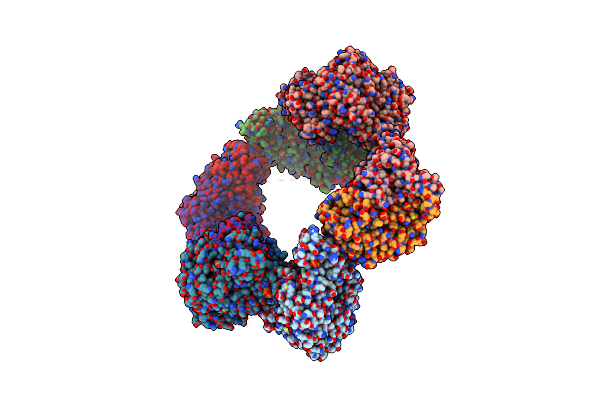 |
Crystal Structure Of The Datp Inhibited E. Coli Class Ia Ribonucleotide Reductase Complex Bound To Cdp And Datp At 2.97 Angstroms Resolution
Organism: Escherichia coli (strain k12)
Method: X-RAY DIFFRACTION Resolution:2.98 Å Release Date: 2016-01-20 Classification: OXIDOREDUCTASE Ligands: CDP, DAT, MG, DTP, FEO |
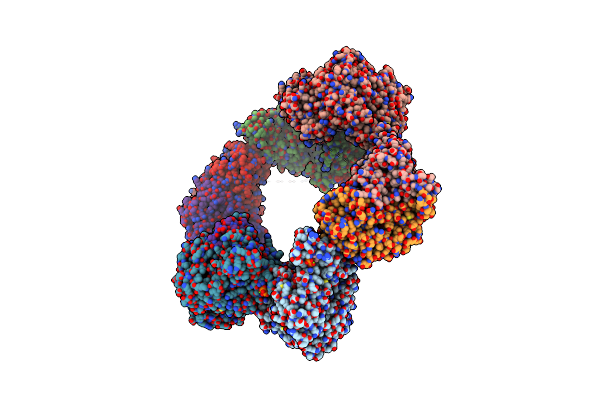 |
Crystal Structure Of The Datp Inhibited E. Coli Class Ia Ribonucleotide Reductase Complex Bound To Adp And Dgtp At 3.40 Angstroms Resolution
Organism: Escherichia coli (strain k12)
Method: X-RAY DIFFRACTION Resolution:3.40 Å Release Date: 2016-01-20 Classification: OXIDOREDUCTASE Ligands: DGT, MG, ADP, DAT, FEO |
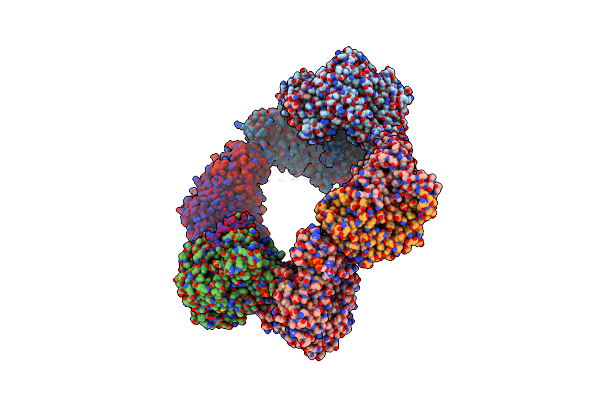 |
Crystal Structure Of The Datp Inhibited E. Coli Class Ia Ribonucleotide Reductase Complex Bound To Gdp And Ttp At 3.20 Angstroms Resolution
Organism: Escherichia coli (strain k12)
Method: X-RAY DIFFRACTION Resolution:3.20 Å Release Date: 2016-01-20 Classification: OXIDOREDUCTASE Ligands: GDP, DAT, MG, TTP, FEO |
 |
Crystallographic Structure Of Nucleoside Diphosphate Kinase From Litopenaeus Vannamei Complexed With Dadp
Organism: Litopenaeus vannamei
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2014-09-10 Classification: TRANSFERASE Ligands: DAT, MG |
 |
Crystal Structure Of The Datp Inhibited E. Coli Class Ia Ribonucleotide Reductase Complex At 4 Angstroms Resolution
Organism: Escherichia coli k-12
Method: X-RAY DIFFRACTION Resolution:3.95 Å Release Date: 2012-07-04 Classification: OXIDOREDUCTASE Ligands: DTP, DAT, MG, FEO |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2010-10-06 Classification: PROTEIN BINDING, TRANSFERASE Ligands: DTV, EPE, DAT, SO4 |
 |
Crystal Structure Of Deletion Mutant Of Aps-Kinase Domain Of Human Paps-Synthetase 1
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.88 Å Release Date: 2007-05-29 Classification: TRANSFERASE Ligands: ADX, DAT |
 |
Crystal Structrue Of Deletion Mutant Of Aps-Kinase Domain Of Human Paps-Synthetase 1 In Complex With Cyclic Paps And Dadp
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.40 Å Release Date: 2007-05-29 Classification: TRANSFERASE Ligands: GGZ, DAT |
 |
Crystal Structures Of The Hslvu Peptidase-Atpase Complex Reveal An Atp-Dependent Proteolysis Mechanism
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:3.00 Å Release Date: 2001-02-21 Classification: CHAPERONE/HYDROLASE Ligands: DAT |

