Search Count: 102
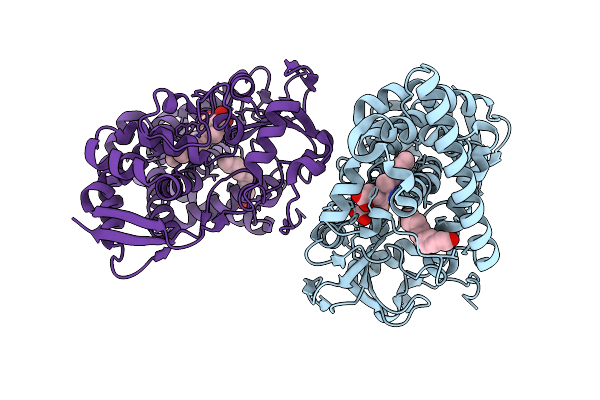 |
Crystal Structure Of The Cyp153A Double Mutant L354V/V456G From Marinobacter Aquaeolei
Organism: Marinobacter nauticus
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2025-11-19 Classification: OXIDOREDUCTASE Ligands: HEM, DAO |
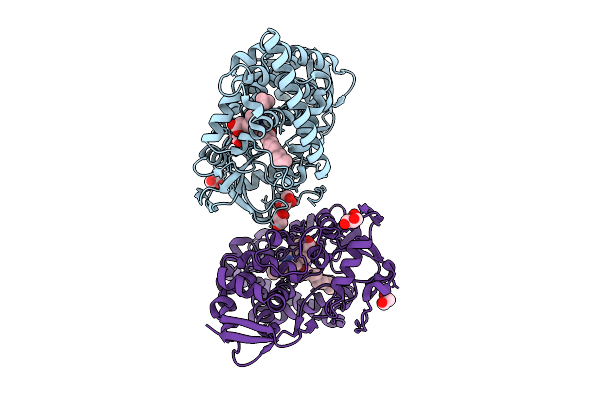 |
Crystal Structure Of The Cyp153A Double Mutant L354T/V456G From Marinobacter Aquaeolei
Organism: Marinobacter nauticus
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2025-08-06 Classification: OXIDOREDUCTASE Ligands: HEM, DAO, GOL |
 |
Unspecific Peroxygenase From Psathyrella Aberdarensis (Pabupo-Ii) In Complex With Lauric Acid
Organism: Candolleomyces aberdarensis
Method: X-RAY DIFFRACTION Resolution:2.15 Å Release Date: 2025-02-26 Classification: OXIDOREDUCTASE Ligands: DAO, EPE, GOL, HEM, SO4, MG |
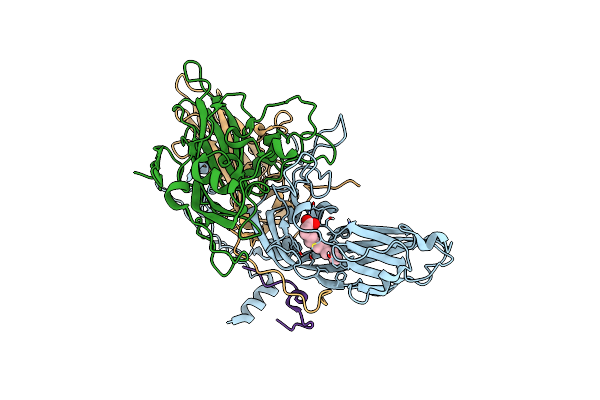 |
Organism: Human rhinovirus 89 atcc vr-1199
Method: ELECTRON MICROSCOPY Release Date: 2024-12-18 Classification: VIRUS Ligands: DAO |
 |
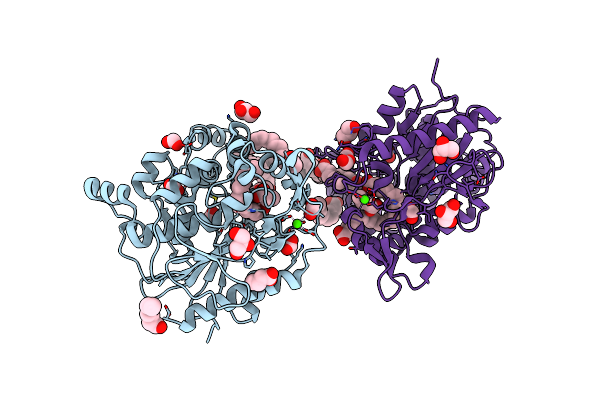
Unspecific Peroxygenase From Marasmius Wettsteinii (Mweupo-1) In Complex With Lauric Acid
Organism: Marasmius wettsteinii
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2024-10-16 Classification: OXIDOREDUCTASE Ligands: NAG, DAO, HEM, PEG, GOL, MG, SO4 |
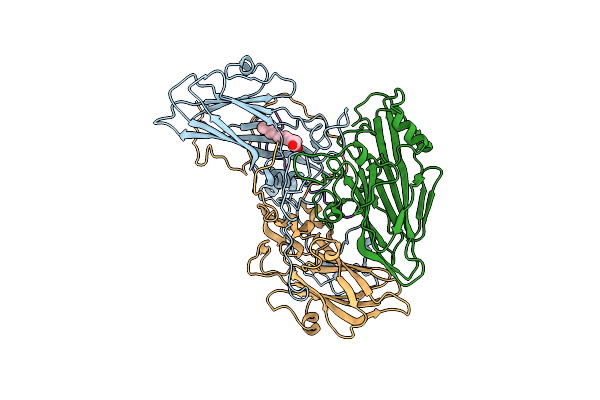 |
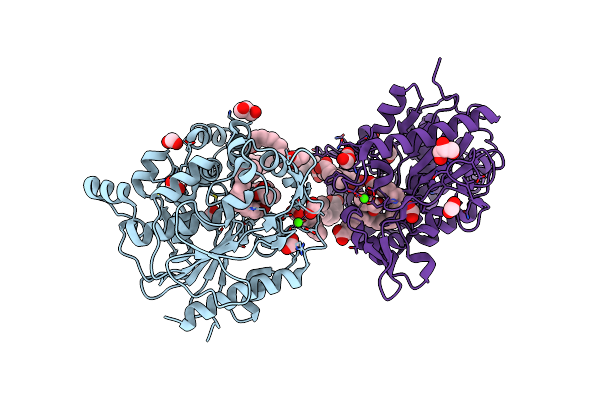
Rhinovirus A2 Uncoating Intermediate Revealing The Natural Pocket Factor (Ph 5.8 And 4 Degrees Celsius)
Organism: Rhinovirus a2
Method: ELECTRON MICROSCOPY Release Date: 2024-07-31 Classification: VIRUS Ligands: DAO |
 |
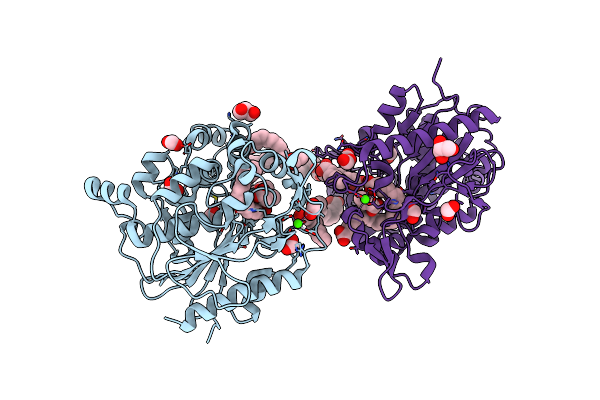
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2024-06-19 Classification: FLAVOPROTEIN Ligands: FAD, DAO |
 |
 |
 |
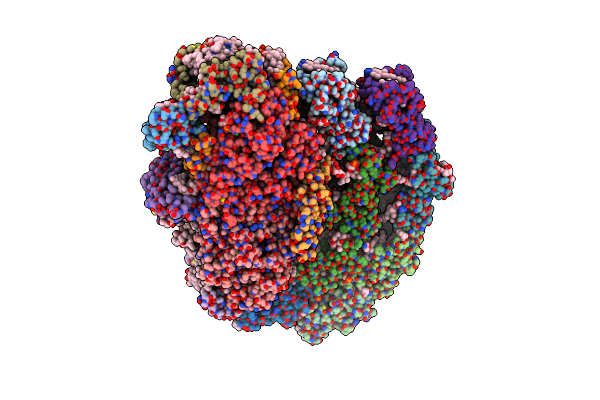 |
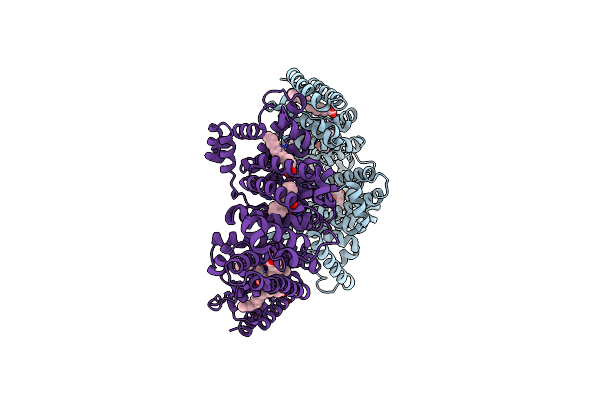
Cryo-Em Structure Of The Photosystem I - Lhci Supercomplex From Coelastrella Sp.
|
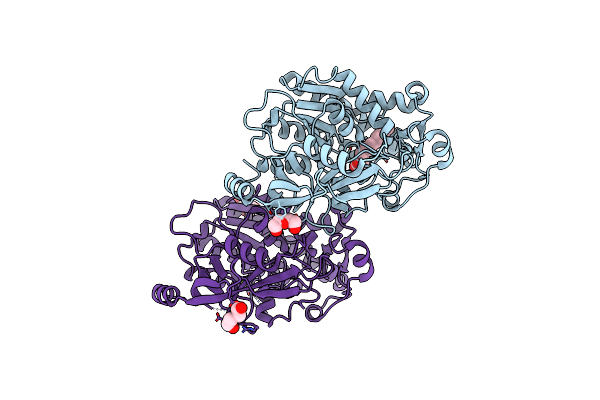 |
Structure Of The Thermolabile Hemolysin From Vibrio Alginolyticus (In Complex With Lauric Acid)
Organism: Vibrio alginolyticus
Method: X-RAY DIFFRACTION Resolution:1.94 Å Release Date: 2023-08-16 Classification: HYDROLASE Ligands: DAO, PEG, MG |
 |
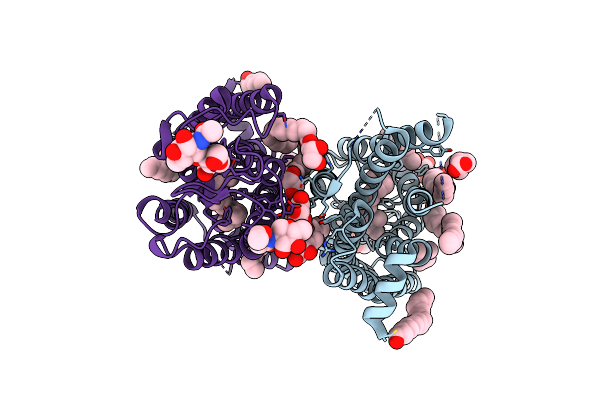
Computational Design Of Stable Mammalian Serum Albumins For Bacterial Expression
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2023-05-10 Classification: TRANSPORT PROTEIN Ligands: MYR, RWF, DAO |
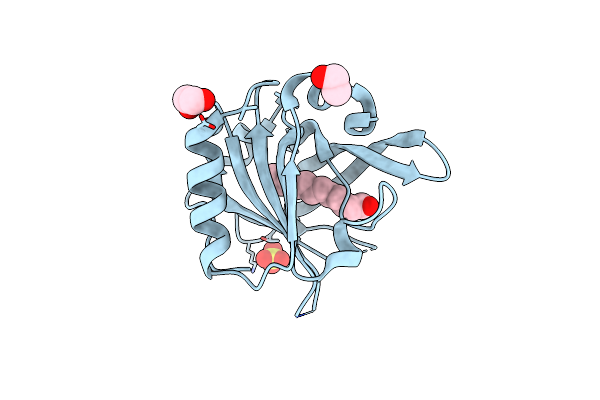 |
Mutant L39Y-L58F Of Recombinant Bovine Beta-Lactoglobulin In Complex With Endogenous Fatty Acid
Organism: Bos taurus
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2023-04-26 Classification: TRANSPORT PROTEIN Ligands: DAO, EDO, SO4, CL |
 |
Dark State Crystal Structure Of Bovine Rhodopsin In Lipidic Cubic Phase (Sacla)
Organism: Bos taurus
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2023-03-29 Classification: MEMBRANE PROTEIN Ligands: ACE, RET, NAG, DAO, OLC, PLM |
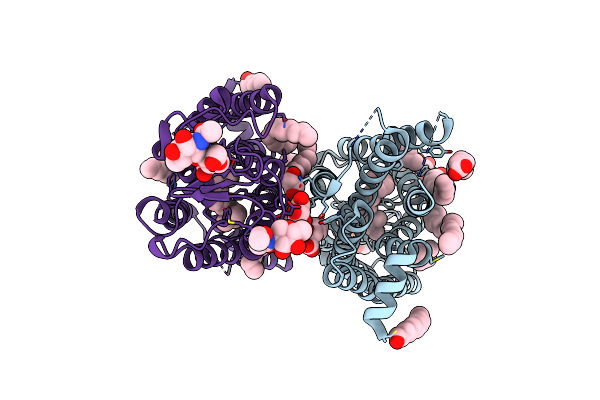 |
Dark State Crystal Structure Of Bovine Rhodopsin In Lipidic Cubic Phase (Swissfel)
Organism: Bos taurus
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2023-03-29 Classification: MEMBRANE PROTEIN Ligands: ACE, RET, NAG, DAO, OLC, PLM |
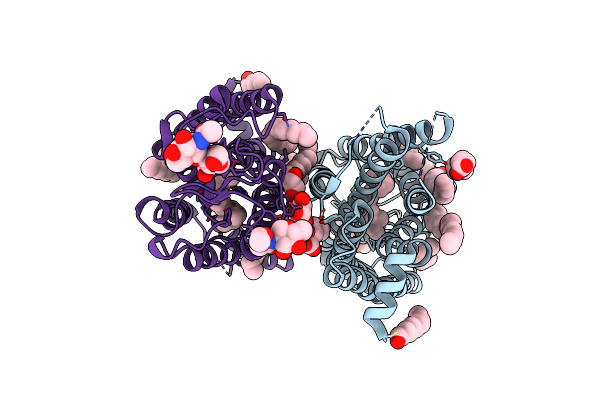 |
1 Picosecond Light Activated Crystal Structure Of Bovine Rhodopsin In Lipidic Cubic Phase
Organism: Bos taurus
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2023-03-29 Classification: MEMBRANE PROTEIN Ligands: ACE, RET, NAG, DAO, OLC, PLM |
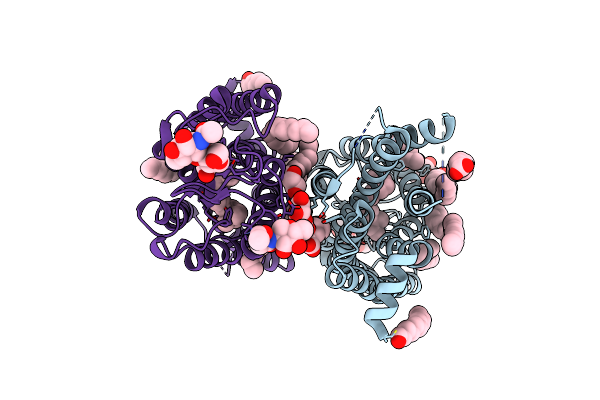 |
10 Picosecond Light Activated Crystal Structure Of Bovine Rhodopsin In Lipidic Cubic Phase
Organism: Bos taurus
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2023-03-29 Classification: MEMBRANE PROTEIN Ligands: ACE, RET, NAG, DAO, OLC, PLM |
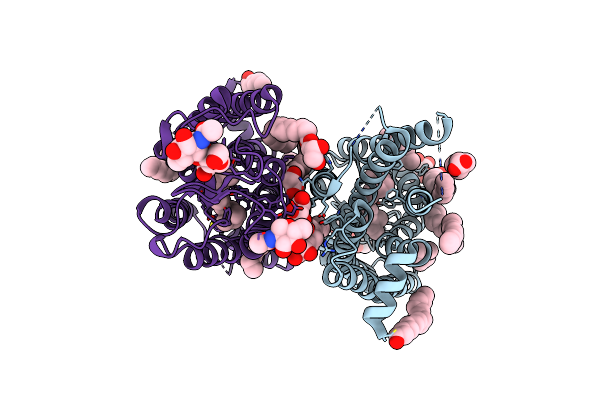 |
100 Picosecond Light Activated Crystal Structure Of Bovine Rhodopsin In Lipidic Cubic Phase (Sacla)
Organism: Bos taurus
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2023-03-29 Classification: MEMBRANE PROTEIN Ligands: RET, NAG, DAO, OLC, PLM |
 |

