Search Count: 47
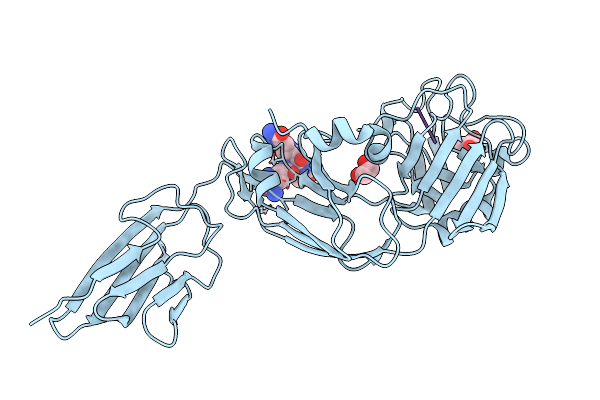 |
Crystal Structure Of The Transpeptidase Ldtmt2 From Mycobacterium Tuberculosis In Complex With Natural Substrate
Organism: Mycobacterium tuberculosis, Corynebacterium jeikeium
Method: X-RAY DIFFRACTION Resolution:1.98 Å Release Date: 2024-08-07 Classification: ANTIMICROBIAL PROTEIN Ligands: EDO, DAL, CL |
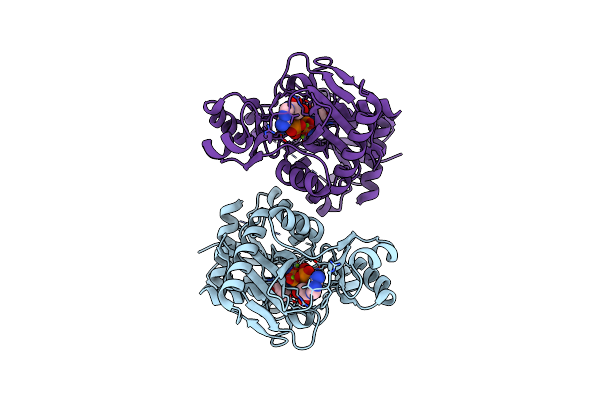 |
Organism: Pseudomonas aeruginosa
Method: X-RAY DIFFRACTION Resolution:1.22 Å Release Date: 2023-08-30 Classification: LIGASE/PRODUCT Ligands: ATP, DAL, MG |
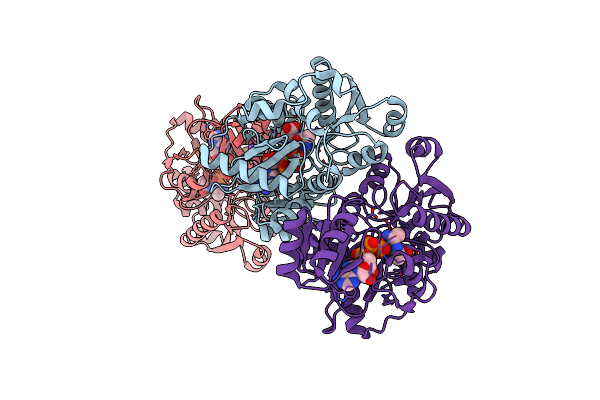 |
Organism: Pseudomonas aeruginosa
Method: X-RAY DIFFRACTION Resolution:2.35 Å Release Date: 2023-08-30 Classification: LIGASE/PRODUCT Ligands: DAL, MG, ATP, NA |
 |
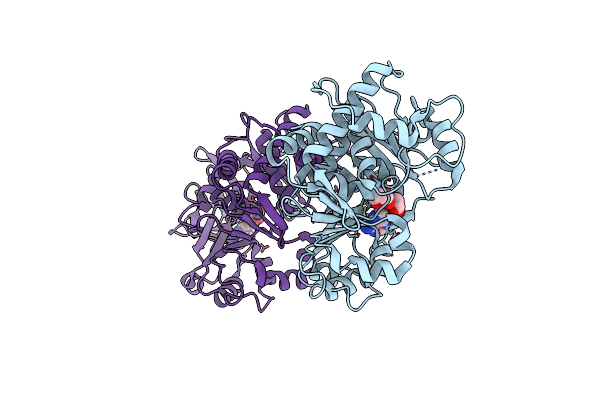
Co-Crystal Structure Of Salmonella Typhimurium Tlde1A In Complex With D-Alanine
Organism: Salmonella enterica subsp. enterica serovar typhimurium, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2022-11-02 Classification: TOXIN Ligands: DAL, SO4 |
 |
Organism: Lactiplantibacillus plantarum subsp. plantarum nc8
Method: X-RAY DIFFRACTION Resolution:2.01 Å Release Date: 2022-08-03 Classification: CYTOSOLIC PROTEIN Ligands: DAL, AMP |
 |
Crystal Structure Of The Peptidoglycan Binding Domain Of The Outer Membrane Protein (Ompa) From Klebsiella Pneumoniae With Bound D-Alanine
Organism: Klebsiella pneumoniae subsp. pneumoniae
Method: X-RAY DIFFRACTION Resolution:1.88 Å Release Date: 2021-07-28 Classification: PEPTIDE BINDING PROTEIN Ligands: DAL, CL |
 |
Organism: Aeromonas hydrophila
Method: X-RAY DIFFRACTION Resolution:2.70 Å Release Date: 2020-07-15 Classification: LIGASE Ligands: DAL |
 |
Thermus Thermophilus D-Alanine-D-Alanine Ligase In Complex With Atp, D-Alanine-D-Alanine, Mg2+ And Rb+
Organism: Thermus thermophilus (strain hb8 / atcc 27634 / dsm 579)
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2020-05-06 Classification: LIGASE Ligands: DAL, MG, RB, ATP |
 |
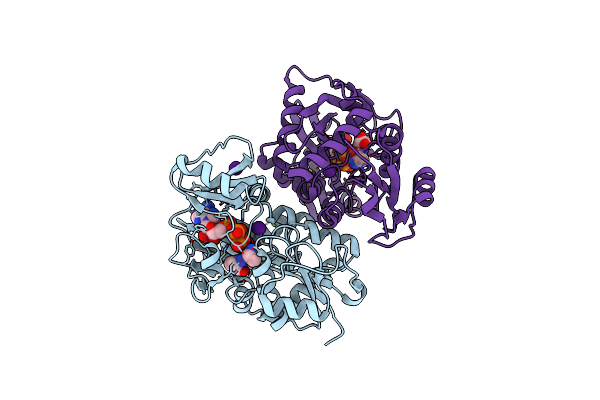
Thermus Thermophilus D-Alanine-D-Alanine Ligase In Complex With Atp, D-Alanine-D-Alanine, Mg2+ And Rb+
Organism: Thermus thermophilus (strain hb8 / atcc 27634 / dsm 579)
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2020-05-06 Classification: LIGASE Ligands: DAL, MG, RB, ATP |
 |
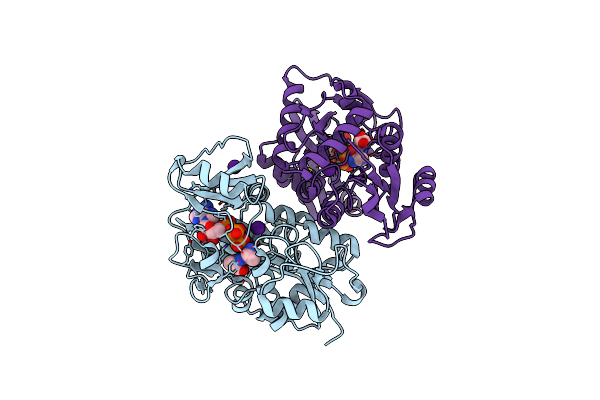
Thermus Thermophilus D-Alanine-D-Alanine Ligase In Complex With Atp, D-Alanine-D-Alanine, Mg2+ And Cs+
Organism: Thermus thermophilus (strain hb8 / atcc 27634 / dsm 579)
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2020-05-06 Classification: LIGASE Ligands: DAL, MG, CS, ATP |
 |
Thermus Thermophilus D-Alanine-D-Alanine Ligase In Complex With Atp, D-Alanine-D-Alanine, Mg2+ And Cs+
Organism: Thermus thermophilus (strain hb8 / atcc 27634 / dsm 579)
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2020-05-06 Classification: LIGASE Ligands: DAL, CS, MG, ATP |
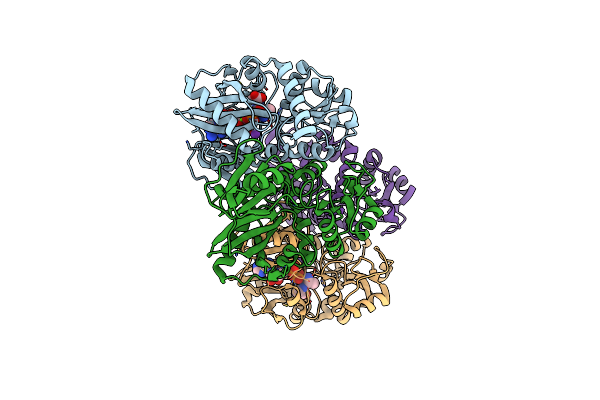 |
Thermus Thermophilus D-Alanine-D-Alanine Ligase In Complex With Adp, Phosphate, D-Ala-D-Ala, Mg2+ And K+
Organism: Thermus thermophilus (strain hb8 / atcc 27634 / dsm 579)
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2020-05-06 Classification: LIGASE Ligands: DAL, MG, PO4, K, ADP |
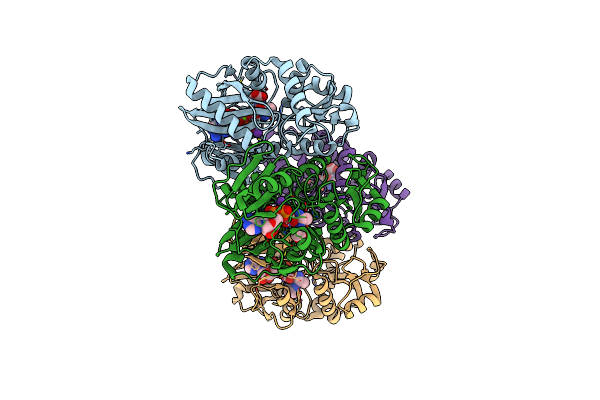 |
Thermus Thermophilus D-Alanine-D-Alanine Ligase In Complex With Adp, Carbonate, D-Alanine-D-Alanine, Mg2+ And K+
Organism: Thermus thermophilus (strain hb8 / atcc 27634 / dsm 579)
Method: X-RAY DIFFRACTION Resolution:1.67 Å Release Date: 2020-05-06 Classification: LIGASE Ligands: DAL, MG, K, ADP, CO3 |
 |
Ompa-Like Domain Of Fopa1 From Francisella Tularensis Subsp. Tularensis Schu S4
Organism: Francisella tularensis subsp. tularensis (strain schu s4 / schu 4)
Method: X-RAY DIFFRACTION Resolution:2.36 Å Release Date: 2019-09-18 Classification: PEPTIDE BINDING PROTEIN Ligands: EPE, 1PE, DAL |
 |
Crystal Structure Of The Human Dual Specificity 1 Catalytic Domain (C258S) As A Maltose Binding Protein Fusion In Complex With The Designed Ar Protein Mbp3_16
Organism: Escherichia coli (strain k12), Homo sapiens, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.23 Å Release Date: 2018-09-19 Classification: HYDROLASE Ligands: PEG, PO4, GLY, PGE, EDO, PG4, DAL |
 |
Organism: Trichodesmium erythraeum ims101
Method: X-RAY DIFFRACTION Resolution:1.10 Å Release Date: 2018-09-12 Classification: METAL BINDING PROTEIN Ligands: DAL, DGL, FE |
 |
Organism: Enterococcus faecalis v583
Method: X-RAY DIFFRACTION Resolution:2.18 Å Release Date: 2018-09-05 Classification: HYDROLASE Ligands: CU, DAL, GOL, PG4 |
 |
Organism: Enterococcus faecalis
Method: X-RAY DIFFRACTION Resolution:2.26 Å Release Date: 2018-09-05 Classification: HYDROLASE Ligands: DAL, GOL, ACT, ZN, PGE |
 |
Crystal Structure Of Burkholderia Pseudomallei D-Alanine-D-Alanine Ligase In Complex With Amp And D-Ala-D-Ala
Organism: Burkholderia pseudomallei
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2018-05-30 Classification: LIGASE Ligands: AMP, DAL, MG, SO4, EDO, PGE |
 |
Crystal Structure Of Streptavidin D-Amino Acid Containing Peptide Gdlwqheatwkkq
Organism: Streptomyces avidinii
Method: X-RAY DIFFRACTION Resolution:1.61 Å Release Date: 2017-10-04 Classification: Biotin binding protein Ligands: DLE, DTR, DGN, DHI, DGL, DAL, DTH, DLY |

