Search Count: 29
 |
Crystal Structure Of P110Alpha-Ras Binding Domain (Rbd) In Complex With Molecular Glue D927
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-07-23 Classification: SIGNALING PROTEIN Ligands: A1ATF, MG, MRD, MPD |
 |
Crystal Structure Of P110Alpha-Ras Binding Domain (Rbd) In Complex With Molecular Glue D223
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-07-23 Classification: SIGNALING PROTEIN Ligands: A1AZD |
 |
Crystal Structure Of The Kras-P110Alpha Complex In The Presence Of Molecular Glue D223
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-07-23 Classification: ONCOPROTEIN Ligands: A1AZD, GOL, MG, GNP |
 |
Crystal Structure Of P110Alpha-Rbd Covalently Bound To A Breaker Compound Bbo-10203 Via Cys242
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-06-25 Classification: ONCOPROTEIN Ligands: A1AIR |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.46 Å Release Date: 2025-01-22 Classification: ONCOPROTEIN Ligands: GNP, MG |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2025-01-22 Classification: ONCOPROTEIN Ligands: GNP, MG, ZN |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:3.10 Å Release Date: 2025-01-22 Classification: ONCOPROTEIN Ligands: 5H5, GNP, MG |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.75 Å Release Date: 2025-01-22 Classification: ONCOPROTEIN Ligands: 5H5, GNP, MG |
 |
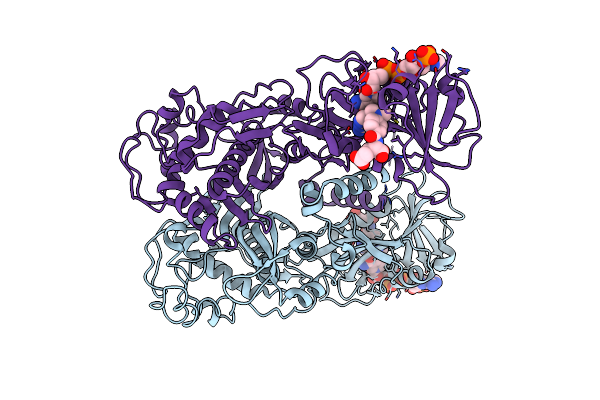
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.81 Å Release Date: 2025-01-22 Classification: ONCOPROTEIN Ligands: A1ATF, MG, IPA, GNP |
 |
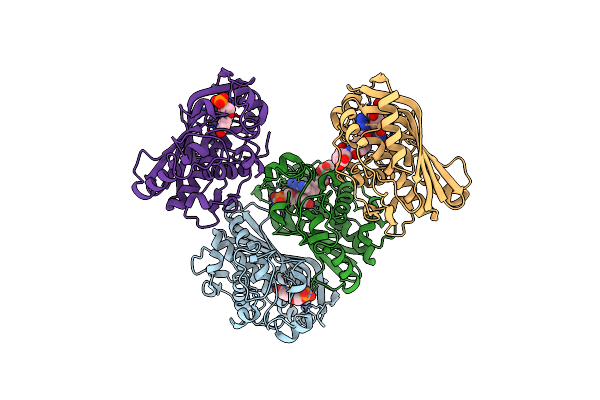
Organism: Cryptosporidium hominis
Method: X-RAY DIFFRACTION Resolution:2.79 Å Release Date: 2020-06-17 Classification: TRANSFERASE Ligands: NDP, MTX |
 |
Crystal Structure Of Ts-Dhfr From Cryptosporidium Hominis In Complex With Nadph, Fdump And 2-(4-((2-Amino-4-Oxo-4,7-Dihydro-3H-Pyrrolo[2,3-D]Pyrimidin-5-Yl)Methyl)Benzamido)Terephthalic Acid
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2019-10-16 Classification: TRANSFERASE/TRANSFERASE INHIBITOR Ligands: UMP, OEJ |
 |
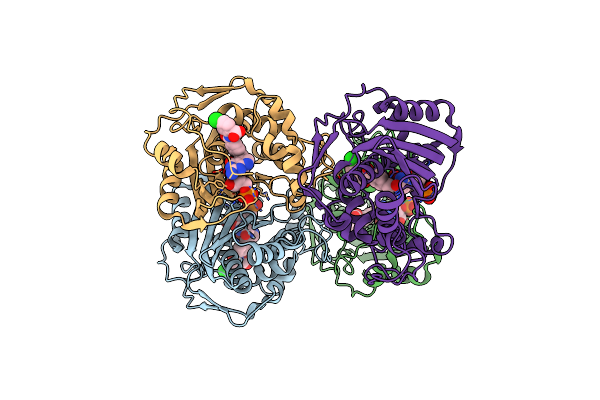
Crystal Structure Of Ts-Dhfr From Cryptosporidium Hominis In Complex With Nadph, Fdump And 2-(2-(4-((2-Amino-4-Oxo-4,7-Dihydro-3H-Pyrrolo[2,3-D]Pyrimidin-5-Yl)Methyl)Benzamido)-4-Methoxyphenyl)Acetic Acid
Organism: Cryptosporidium hominis
Method: X-RAY DIFFRACTION Resolution:2.69 Å Release Date: 2019-10-16 Classification: TRANSFERASE/TRANSFERASE INHIBITOR Ligands: NDP, UFP, OED, MTX |
 |
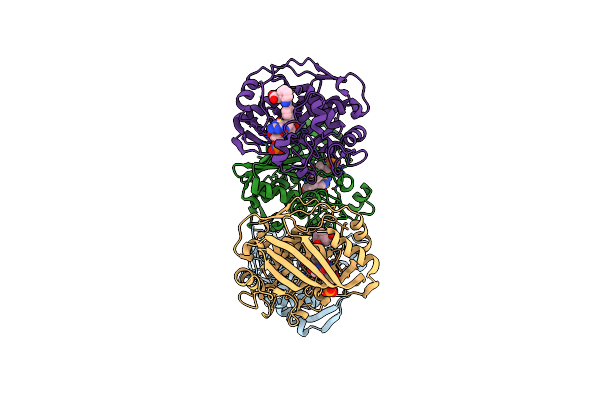
Crystal Structure Of Human Thymidylate Synthase Delta (7-29) In Complex With Dump And 2-(4-((2-Amino-4-Oxo-4,7-Dihydro-3H-Pyrrolo[2,3-D]Pyrimidin-5-Yl)Methyl)Benzamido)-4-Chlorobenzoic Acid
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.39 Å Release Date: 2019-10-02 Classification: transferase/transferase inhibitor Ligands: UMP, OE7 |
 |
Crystal Structure Of Human Thymidylate Synthase Delta (7-29) In Complex With Dump And 2-(2-(4-((2-Amino-4-Oxo-4,7-Dihydro-3H-Pyrrolo[2,3-D]Pyrimidin-5-Yl)Methyl)Benzamido)Phenyl)Acetic Acid
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.85 Å Release Date: 2019-10-02 Classification: transferase/transferase inhibitor Ligands: UMP, OF7 |
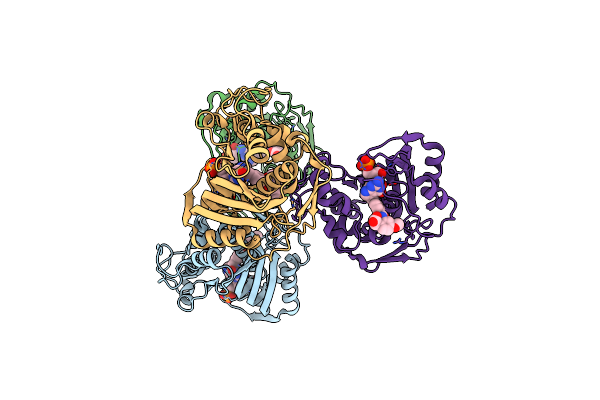 |
Crystal Structure Of Human Thymidylate Synthase Delta (7-29) In Complex With Dump And 2-(2-(4-((2-Amino-4-Oxo-4,7-Dihydro-3H-Pyrrolo[2,3-D]Pyrimidin-5-Yl)Methyl)Benzamido)-4-Methoxyphenyl)Acetic Acid
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.39 Å Release Date: 2019-10-02 Classification: transferase/transferase inhibitor Ligands: UMP, OED |
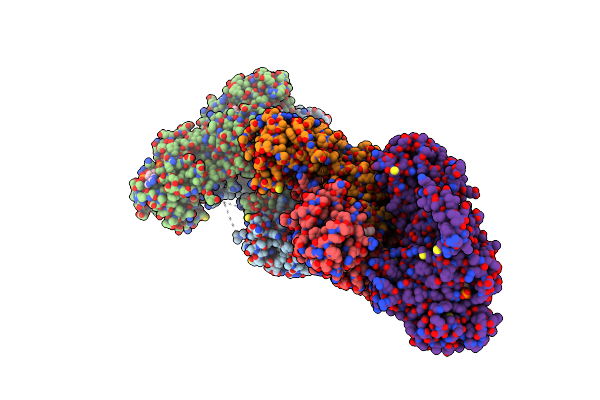 |
Crystal Structure Of Ts-Dhfr From Cryptosporidium Hominis In Complex With Nadph, Fdump And 2-(4-((2-Amino-4-Oxo-4,7-Dihydro-3H-Pyrrolo[2,3-D]Pyrimidin-5-Yl)Methyl)Benzamido)Benzoic Acid
Organism: Cryptosporidium hominis
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2019-10-02 Classification: TRANSFERASE/TRANSFERASE INHIBITOR Ligands: NDP, UFP, OG7, MTX, SO4 |
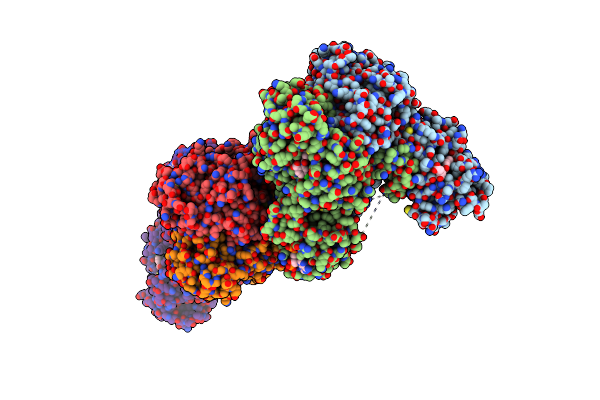 |
Crystal Structure Of Ts-Dhfr From Cryptosporidium Hominis In Complex With Nadph, Fdump And 2-(4-((2-Amino-4-Oxo-4,7-Dihydro-3H-Pyrrolo[2,3-D]Pyrimidin-5-Yl)Methyl)Benzamido)-4-Chlorobenzoic Acid
Organism: Cryptosporidium hominis
Method: X-RAY DIFFRACTION Resolution:2.53 Å Release Date: 2019-10-02 Classification: TRANSFERASE/TRANSFERASE INHIBITOR Ligands: NDP, UFP, OE7, MTX |
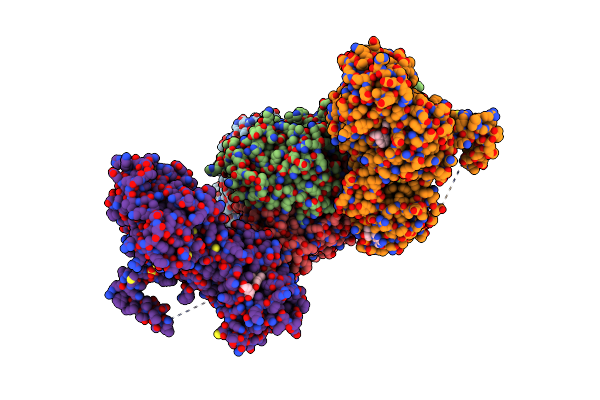 |
Crystal Structure Of Ts-Dhfr From Cryptosporidium Hominis In Complex With Nadph, Fdump And 2-(2-(4-((2-Amino-4-Oxo-4,7-Dihydro-3H-Pyrrolo[2,3-D]Pyrimidin-5-Yl)Methyl)Benzamido)Phenyl)Acetic Acid.
Organism: Cryptosporidium hominis
Method: X-RAY DIFFRACTION Resolution:2.89 Å Release Date: 2019-10-02 Classification: TRANSFERASE/TRANSFERASE INHIBITOR Ligands: NDP, UFP, OF7, MTX, SO4 |
 |
Crystal Structure Of Ts-Dhfr From Cryptosporidium Hominis In Complex With Nadph, Fdump And 2-((4-((2-Amino-4-Oxo-4,7-Dihydro-3H-Pyrrolo[2,3-D]Pyrimidin-5-Yl)Methyl)Benzamido)Methyl)Benzoic Acid.
Organism: Cryptosporidium hominis
Method: X-RAY DIFFRACTION Resolution:2.79 Å Release Date: 2019-10-02 Classification: TRANSFERASE/TRANSFERASE INHIBITOR Ligands: NDP, UFP, OF4, MTX, SO4 |
 |
Crystal Structure Of Ts-Dhfr From Cryptosporidium Hominis In Complex With Nadph, Fdump And 3-(2-(4-((2-Amino-4-Oxo-4,7-Dihydro-3H-Pyrrolo[2,3-D]Pyrimidin-5-Yl)Methyl)Benzamido)Phenyl)Propanoic Acid.
Organism: Cryptosporidium hominis
Method: X-RAY DIFFRACTION Resolution:3.09 Å Release Date: 2019-10-02 Classification: TRANSFERASE/TRANSFERASE INHIBITOR Ligands: NDP, UFP, OG4, MTX |

