Search Count: 19
 |
Organism: Saccharomyces cerevisiae, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2025-06-18 Classification: DNA BINDING PROTEIN |
 |
Organism: Saccharomyces cerevisiae, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2025-06-18 Classification: DNA BINDING PROTEIN |
 |
Solution Structure Of The N-Terminal Domain Of Human Telomeric Repeat-Binding Factor 2-Interacting Protein 1 (Hrap1): Implication For Rap1-Trf2 Interaction In Human.
|
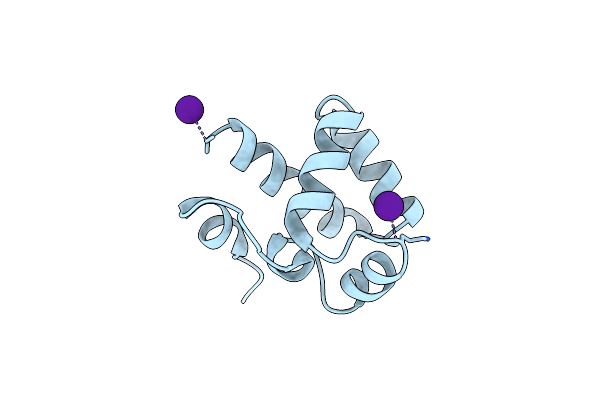 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:3.00 Å Release Date: 2021-07-07 Classification: DNA BINDING PROTEIN Ligands: CS |
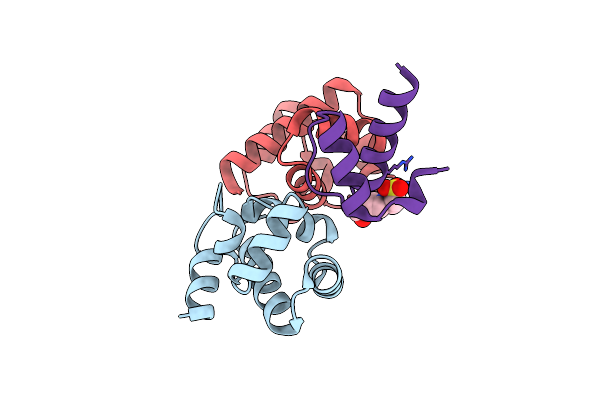 |
Di-Phosphorylated Barrier-To-Autointegration Factor (Baf) In Complex With Lem Domain Of Emerin
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.44 Å Release Date: 2021-04-07 Classification: DNA BINDING PROTEIN Ligands: EPE |
 |
The Ctd Of Hpdpra, A Dna Binding Winged Helix Domain Which Do Not Bind Dsdna
Organism: Helicobacter pylori
Method: SOLUTION NMR Release Date: 2019-04-24 Classification: DNA BINDING PROTEIN |
 |
Organism: Helicobacter pylori 26695
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2017-12-20 Classification: DNA BINDING PROTEIN Ligands: EDO |
 |
Nmr Structure Of The C-Terminal Domain Of The Bacteriophage T5 Decoration Protein Pb10.
Organism: Escherichia phage t5
Method: SOLUTION NMR Release Date: 2017-08-02 Classification: VIRAL PROTEIN |
 |
Nmr Structure Of The N-Terminal Domain Of The Bacteriophage T5 Decoration Protein Pb10
Organism: Escherichia phage t5
Method: SOLUTION NMR Release Date: 2017-04-19 Classification: VIRAL PROTEIN |
 |
Organism: Escherichia phage t5
Method: ELECTRON MICROSCOPY Release Date: 2017-02-22 Classification: VIRAL PROTEIN |
 |
Organism: Mus musculus
Method: X-RAY DIFFRACTION Resolution:2.60 Å Release Date: 2014-12-17 Classification: HYDROLASE |
 |
Organism: Mus musculus, Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.94 Å Release Date: 2014-12-17 Classification: IMMUNE SYSTEM Ligands: CL, EPE, EDO, ACT, MG |
 |
Organism: Mus musculus, Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.95 Å Release Date: 2014-12-17 Classification: IMMUNE SYSTEM Ligands: CL, GOL, EPE, EDO, MG |
 |
Novel Inhibition Mechanism Of Membrane Metalloprotease By An Exosite-Swiveling Conformational Antibody
Organism: Mus musculus, Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2014-12-17 Classification: IMMUNE SYSTEM Ligands: SO4 |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.85 Å Release Date: 2013-08-21 Classification: HYDROLASE Ligands: ZN, CA, CL, EDO |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.44 Å Release Date: 2013-08-21 Classification: HYDROLASE Ligands: ZN, CA, CL, GOL, PGO, PEG |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.54 Å Release Date: 2013-08-21 Classification: HYDROLASE Ligands: ZN, CA, CL, PGO, PEG, GOL |
 |
General Structure-Based Approach To The Design Of Protein Ligands: Application To The Design Of Kv1.2 Potassium Channel Blockers.
Organism: Mus musculus
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2006-11-21 Classification: Immune System/DNA |
 |
Crystal Structure Of The Stromelysin-3 (Mmp-11) Catalytic Domain Complexed With A Phosphinic Inhibitor
Organism: Mus musculus
Method: X-RAY DIFFRACTION Resolution:2.60 Å Release Date: 2001-03-28 Classification: HYDROLASE Ligands: ZN, CA, CPS, RXP |

