Search Count: 70
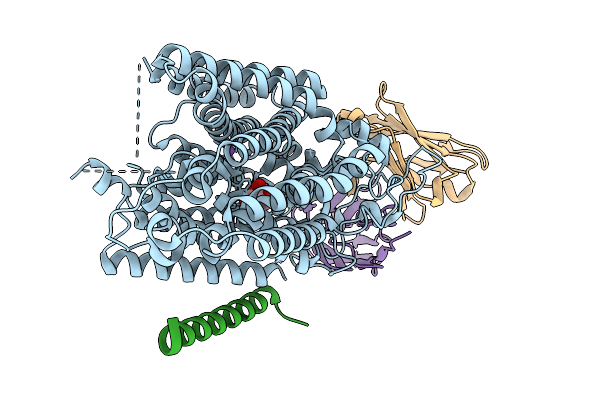 |
Structure Of Hsglt2-Map17 Complex In The Substrate-Bound Occluded Conformation
Organism: Homo sapiens, Mus musculus
Method: ELECTRON MICROSCOPY Release Date: 2025-09-03 Classification: TRANSPORT PROTEIN |
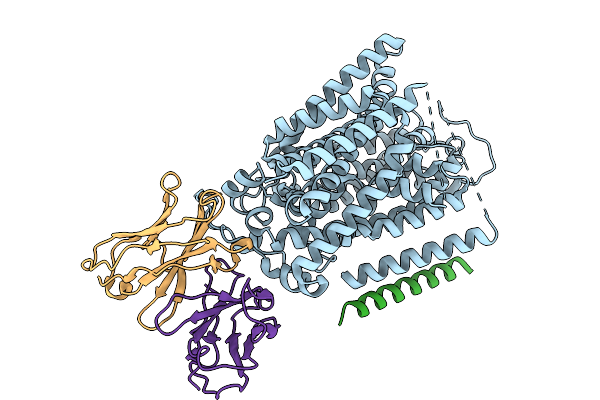 |
Structure Of Hsglt2-Map17 Complex In The Substrate-Free Inward-Facing Conformation In The Presence Of Sodium
Organism: Homo sapiens, Mus musculus
Method: ELECTRON MICROSCOPY Release Date: 2025-09-03 Classification: TRANSPORT PROTEIN |
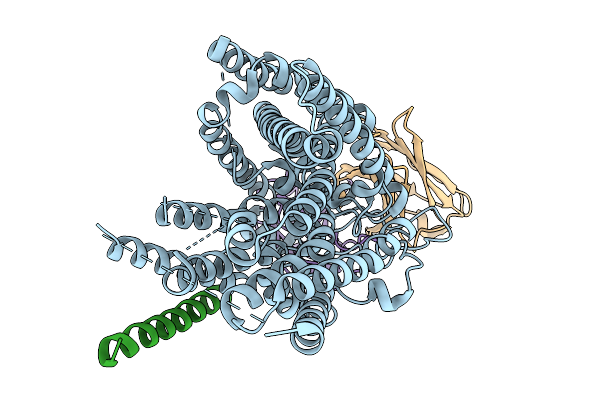 |
Structure Of Hsglt2-Map17 Complex In The Substrate-Free Inward-Facing Conformation In The Presence Of Potassium
Organism: Homo sapiens, Mus musculus
Method: ELECTRON MICROSCOPY Release Date: 2025-09-03 Classification: TRANSPORT PROTEIN |
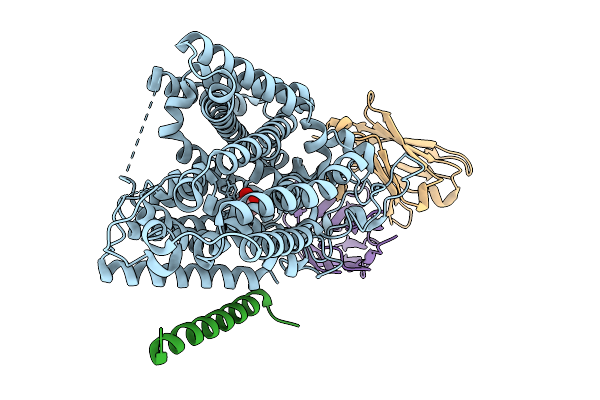 |
Structure Of Hsglt2-Map17 Complex In The Substrate-Bound Inward-Facing Conformation
Organism: Homo sapiens, Mus musculus
Method: ELECTRON MICROSCOPY Release Date: 2025-09-03 Classification: TRANSPORT PROTEIN Ligands: A1EFW |
 |
Organism: Rattus norvegicus
Method: ELECTRON MICROSCOPY Release Date: 2025-07-23 Classification: MEMBRANE PROTEIN Ligands: A1D92 |
 |
Organism: Monkeypox virus
Method: X-RAY DIFFRACTION Resolution:2.83 Å Release Date: 2025-04-16 Classification: VIRAL PROTEIN Ligands: ZN, PO4, TTP |
 |
Organism: Monkeypox virus, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2025-03-12 Classification: VIRAL PROTEIN |
 |
Organism: Monkeypox virus, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2025-03-12 Classification: VIRAL PROTEIN |
 |
Organism: Monkeypox virus, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2025-03-12 Classification: VIRAL PROTEIN |
 |
Organism: Monkeypox virus
Method: ELECTRON MICROSCOPY Release Date: 2025-03-12 Classification: VIRAL PROTEIN |
 |
Organism: Monkeypox virus
Method: ELECTRON MICROSCOPY Release Date: 2025-03-12 Classification: VIRAL PROTEIN Ligands: E6D |
 |
Cryo-Em Structure Of Mpxv Core Protease In Complex With The Substrate Derivative I-G18
Organism: Monkeypox virus
Method: ELECTRON MICROSCOPY Release Date: 2025-03-12 Classification: VIRAL PROTEIN |
 |
Organism: Taterapox virus
Method: X-RAY DIFFRACTION Release Date: 2025-03-12 Classification: VIRAL PROTEIN |
 |
Organism: Monkeypox virus
Method: X-RAY DIFFRACTION Release Date: 2025-03-12 Classification: VIRAL PROTEIN |
 |
Organism: Mus musculus
Method: ELECTRON MICROSCOPY Release Date: 2024-07-10 Classification: MEMBRANE PROTEIN |
 |
Organism: Mus musculus
Method: ELECTRON MICROSCOPY Release Date: 2024-07-10 Classification: MEMBRANE PROTEIN |
 |
Organism: Mus musculus
Method: ELECTRON MICROSCOPY Release Date: 2024-07-10 Classification: MEMBRANE PROTEIN Ligands: SRO |
 |
Organism: Mus musculus
Method: ELECTRON MICROSCOPY Release Date: 2024-07-10 Classification: MEMBRANE PROTEIN Ligands: SRO |
 |
Organism: Cellulosimicrobium
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2024-04-10 Classification: HYDROLASE Ligands: CA |
 |
Organism: Cellulosimicrobium
Method: X-RAY DIFFRACTION Resolution:2.02 Å Release Date: 2024-04-10 Classification: HYDROLASE Ligands: GOL, CA |

