Search Count: 122
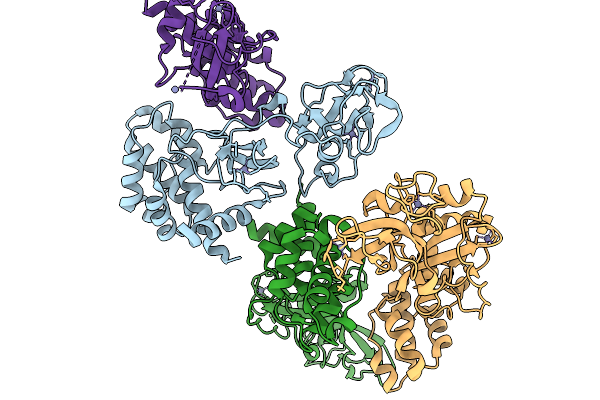 |
Crystal Structure Of Human Shiftless (Sfl) Containing Phosphorylation Sites Ser249, Thr250, Thr253 And Ser256
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-11-19 Classification: ANTIVIRAL PROTEIN Ligands: ZN |
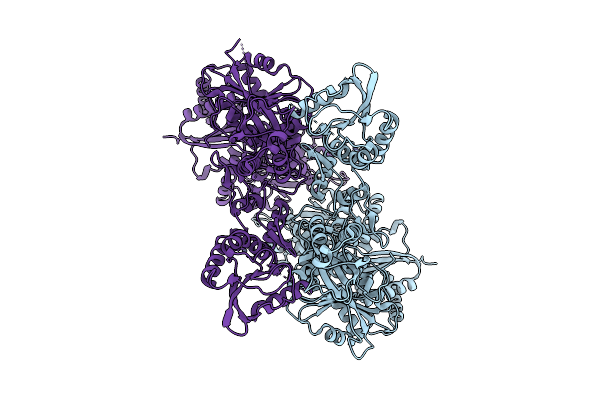 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-08-06 Classification: RNA BINDING PROTEIN Ligands: ZN |
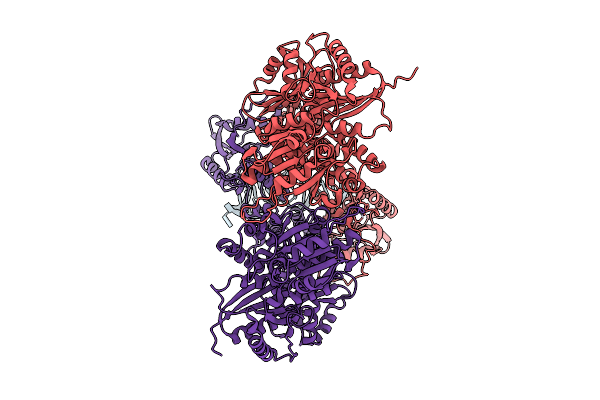 |
Electronic Microscopy Structure Of Human Schlafen14-E211A Dimer In Complex With Dsrna
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-08-06 Classification: RNA BINDING PROTEIN/RNA Ligands: ZN |
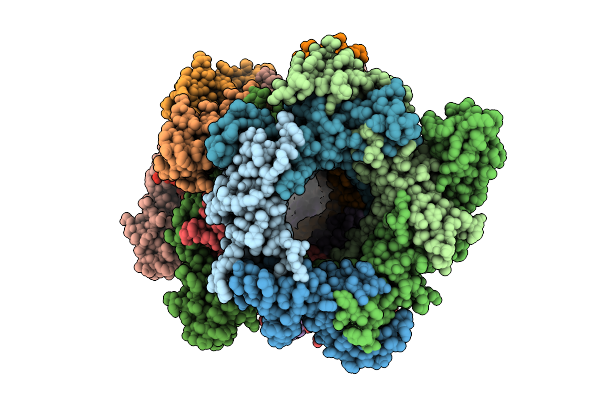 |
Organism: Bacillus subtilis
Method: ELECTRON MICROSCOPY Release Date: 2025-07-16 Classification: ANTIVIRAL PROTEIN |
 |
Organism: Bacillus subtilis
Method: ELECTRON MICROSCOPY Release Date: 2025-07-16 Classification: ANTIVIRAL PROTEIN |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-06-18 Classification: ONCOPROTEIN |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-06-18 Classification: ONCOPROTEIN/INHIBITOR |
 |
Organism: Bacillus subtilis a29
Method: ELECTRON MICROSCOPY Release Date: 2025-03-05 Classification: HYDROLASE |
 |
Organism: Bacillus subtilis a29
Method: ELECTRON MICROSCOPY Release Date: 2025-03-05 Classification: HYDROLASE |
 |
Organism: Bacillus subtilis a29
Method: ELECTRON MICROSCOPY Release Date: 2025-03-05 Classification: HYDROLASE |
 |
Organism: Bacillus subtilis a29
Method: ELECTRON MICROSCOPY Release Date: 2025-03-05 Classification: HYDROLASE |
 |
Organism: Bacillus subtilis a29
Method: ELECTRON MICROSCOPY Release Date: 2025-03-05 Classification: HYDROLASE |
 |
Organism: Bacillus subtilis a29
Method: ELECTRON MICROSCOPY Release Date: 2025-03-05 Classification: HYDROLASE |
 |
Organism: Bacillus subtilis a29
Method: ELECTRON MICROSCOPY Release Date: 2025-03-05 Classification: HYDROLASE |
 |
Organism: Bacillus subtilis a29
Method: ELECTRON MICROSCOPY Release Date: 2025-03-05 Classification: HYDROLASE |
 |
Organism: Bacillus subtilis a29
Method: ELECTRON MICROSCOPY Release Date: 2025-03-05 Classification: HYDROLASE |
 |
Organism: Candidatus accumulibacter adiacens, Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2024-08-28 Classification: VIRAL PROTEIN/HYDROLASE |
 |
Organism: Candidatus accumulibacter adiacens, Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2024-08-28 Classification: VIRAL PROTEIN/HYDROLASE Ligands: NAG |
 |
Organism: Geobacter sulfurreducens, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2024-07-03 Classification: DNA/RNA/CELL INVASION Ligands: MG |
 |
Organism: Geobacter sulfurreducens, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2024-07-03 Classification: DNA/RNA/CELL INVASION Ligands: MG |

