Search Count: 241
 |
Organism: Influenza a virus, Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.06 Å Release Date: 2025-12-10 Classification: VIRAL PROTEIN Ligands: NAG, NA, PEG, EDO, GOL |
 |
Crystal Structure Of Human Cd22 Ig Domains 1-3 In Complex With Modified Sialoside 7-012
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:3.15 Å Release Date: 2025-12-10 Classification: IMMUNE SYSTEM Ligands: A1JJO, SO4 |
 |
Crystal Structure Of Human Cd22 Ig Domains 1-3 In Complex With Modified Sialoside 1B
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.70 Å Release Date: 2025-12-10 Classification: IMMUNE SYSTEM Ligands: A1JJN, GOL |
 |
Organism: Triticum aestivum
Method: ELECTRON MICROSCOPY Resolution:3.33 Å Release Date: 2025-12-10 Classification: PLANT PROTEIN |
 |
Cryo-Em Structure Of Broad Betacoronavirus Binding Antibody 1871 In Complex With Oc43 S2 Subunit
Organism: Human coronavirus oc43, Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-11-12 Classification: VIRAL PROTEIN/IMMUNE SYSTEM |
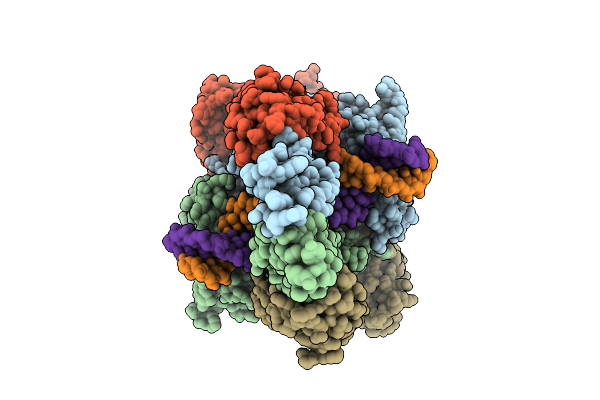 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-10-01 Classification: HYDROLASE Ligands: MG, ADP, BEF, MN |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-10-01 Classification: HYDROLASE Ligands: MG, ADP, BEF, MN |
 |
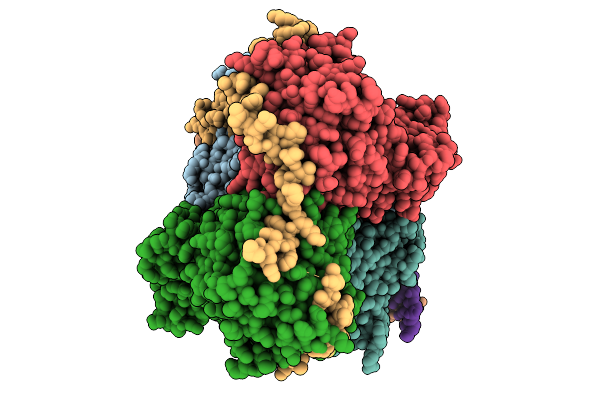
Cryo-Em Structure Of Human Mre11-Rad50-Nbs1 (Mrn) Complex Bound To Dna And Telomeric Factor Trf2 Fragment (438-542)
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-10-01 Classification: HYDROLASE Ligands: MG, ADP, BEF, MN |
 |
Cryo-Em Structure Of Human Mre11-Rad50 (Mr) Complex Bound To Dna And Telomeric Factor Trf2 Fragment (438-542)
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-10-01 Classification: HYDROLASE Ligands: ADP, MG, BEF, MN |
 |
Cryo-Em Structure Of Human Mre11-Rad50-Nbs1 (Mrn) Complex Bound To Dna And Telomeric Factor Trf2
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-10-01 Classification: HYDROLASE Ligands: MG, ADP, BEF, MN |
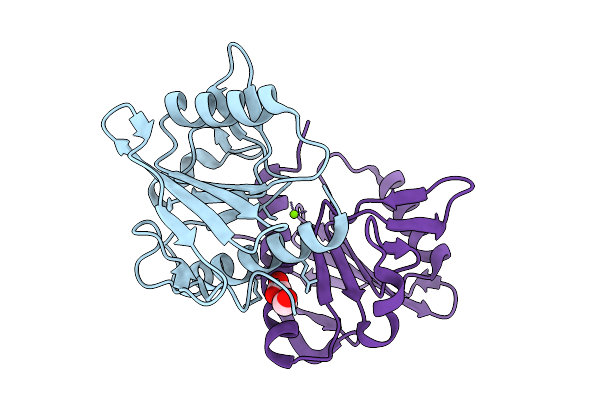 |
Crystal Structure Of Putative Restriction Endonuclease Domain-Containing Protein From Leptospirillum Ferriphilum Ysk
Organism: Leptospirillum ferriphilum ysk
Method: X-RAY DIFFRACTION Resolution:1.65 Å Release Date: 2025-09-10 Classification: HYDROLASE Ligands: FMT, MG, CL |
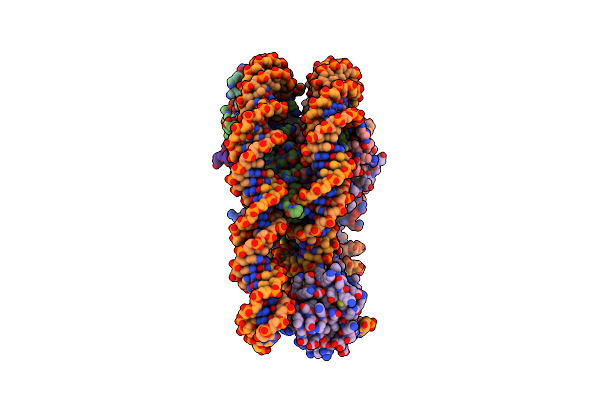 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Resolution:2.69 Å Release Date: 2025-05-07 Classification: GENE REGULATION Ligands: UNX |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Resolution:3.00 Å Release Date: 2025-05-07 Classification: GENE REGULATION Ligands: UNX |
 |
Organism: Influenza a virus, Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2025-03-19 Classification: VIRAL PROTEIN/IMMUNE SYSTEM Ligands: NAG, EDO, PEG, K |
 |
Organism: Influenza a virus, Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.06 Å Release Date: 2025-03-19 Classification: VIRAL PROTEIN/IMMUNE SYSTEM Ligands: NAG, EDO |
 |
Organism: Bacillus subtilis
Method: X-RAY DIFFRACTION Resolution:1.83 Å Release Date: 2024-06-19 Classification: ANTIVIRAL PROTEIN |
 |
Organism: Saccharomyces cerevisiae, Xenopus laevis
Method: ELECTRON MICROSCOPY Release Date: 2023-09-27 Classification: GENE REGULATION Ligands: ZN |
 |
Cryo-Em Structure Of The Histone Deacetylase Complex Rpd3S In Complex With Nucleosome
Organism: Xenopus laevis, Saccharomyces cerevisiae, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2023-09-27 Classification: GENE REGULATION Ligands: ZN |
 |
Organism: Xenopus laevis, Saccharomyces cerevisiae, Synthetic construct
Method: ELECTRON MICROSCOPY Resolution:3.40 Å Release Date: 2023-09-27 Classification: GENE REGULATION |
 |
Composite Cryo-Em Structure Of The Histone Deacetylase Complex Rpd3S In Complex With Nucleosome
Organism: Xenopus laevis, Saccharomyces cerevisiae, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2023-09-27 Classification: GENE REGULATION Ligands: ZN |

