Search Count: 121
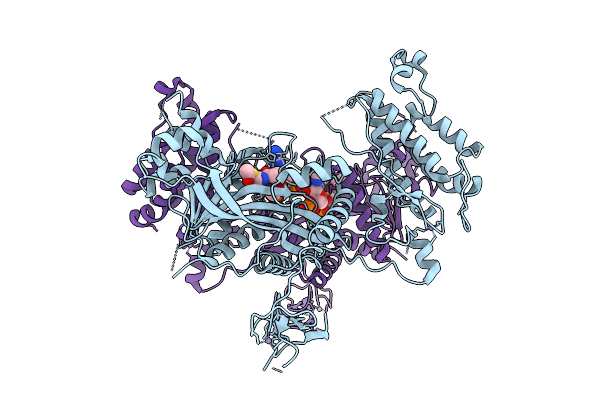 |
Crystal Structure Of Crebbp Histone Acetyltransferase Domain In Complex With Acetyl-Coenzyme A
Organism: Mus musculus
Method: X-RAY DIFFRACTION Release Date: 2025-12-17 Classification: TRANSFERASE Ligands: ZN, ACO |
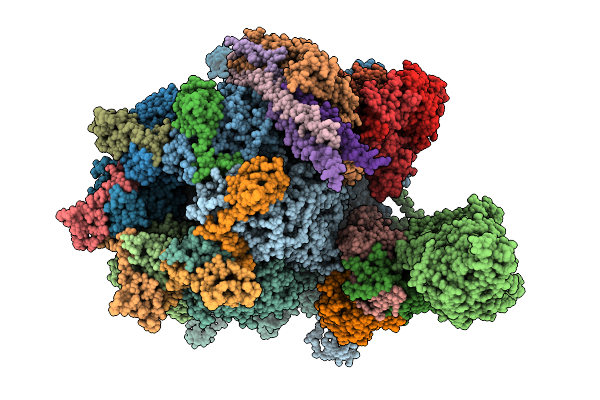 |
Organism: Human alphaherpesvirus 3
Method: ELECTRON MICROSCOPY Release Date: 2025-11-19 Classification: VIRUS |
 |
Crystal Structure Of Human Crebbp Histone Acetyltransferase Domain In Complex With Propionyl- Coenzyme A
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-10-29 Classification: TRANSFERASE Ligands: 1VU, SO4 |
 |
Crystal Structure Of Human Crebbp Histone Acetyltransferase Domain In Complex With A Bisubstrate Inhibitor, Lys-Coa
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-10-15 Classification: TRANSFERASE Ligands: 01K, EDO |
 |
Cryo-Em Structure Of Saccharomyces Cerevisiae Mitochondrial Respiratory Complex Ii
Organism: Saccharomyces cerevisiae
Method: ELECTRON MICROSCOPY Release Date: 2025-09-03 Classification: MEMBRANE PROTEIN Ligands: FAD, FES, SF4, F3S, PEE |
 |
Cryo-Em Structure Of Saccharomyces Cerevisiae Mitochondrial Respiratory Complex Ii In Uq1-Bound State
Organism: Saccharomyces cerevisiae
Method: ELECTRON MICROSCOPY Release Date: 2025-09-03 Classification: MEMBRANE PROTEIN Ligands: FAD, FES, SF4, F3S, UQ1, PEE |
 |
Cryo-Em Structure Of Saccharomyces Cerevisiae Mitochondrial Respiratory Complex Ii In Pydiflumetofen-Bound State
Organism: Saccharomyces cerevisiae
Method: ELECTRON MICROSCOPY Release Date: 2025-09-03 Classification: MEMBRANE PROTEIN Ligands: FAD, FES, SF4, F3S, A1EE4, PEE |
 |
Cryo-Em Structure Of Saccharomyces Cerevisiae Mitochondrial Respiratory Complex Ii In Y19315-Bound State
Organism: Saccharomyces cerevisiae
Method: ELECTRON MICROSCOPY Release Date: 2025-09-03 Classification: MEMBRANE PROTEIN Ligands: FAD, FES, SF4, F3S, A1EKV, PEE |
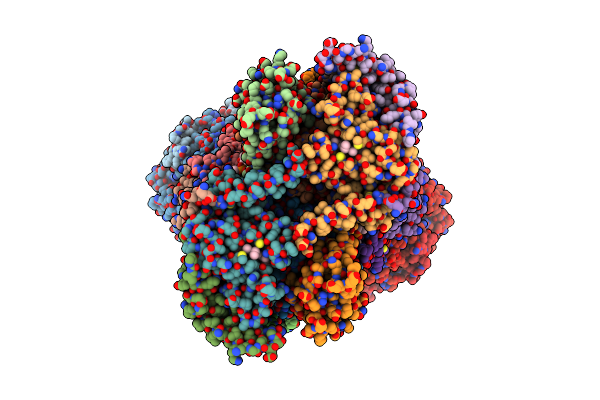 |
Organism: Arachis hypogaea
Method: ELECTRON MICROSCOPY Release Date: 2025-06-04 Classification: PLANT PROTEIN Ligands: ZN, CDL, A1D6K, HEM, 3PE, HEC, FES, PC1 |
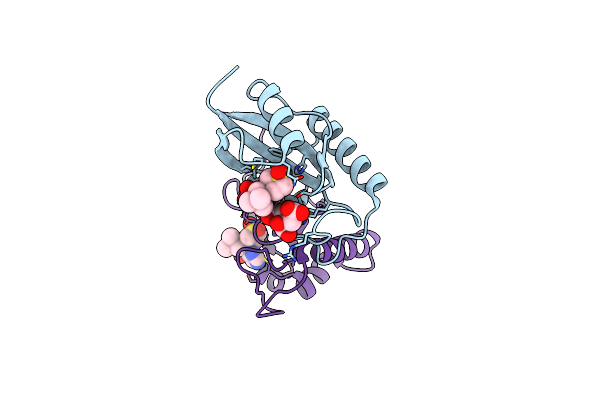 |
Human Pin1 (Peptidyl-Prolyl Cis-Trans Isomerase) Catalytic Domain In Complex With A Covalent Inhibitor
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.57 Å Release Date: 2025-05-14 Classification: ISOMERASE Ligands: A1D9X, CIT |
 |
Organism: Human alphaherpesvirus 3
Method: ELECTRON MICROSCOPY Release Date: 2025-04-30 Classification: VIRAL PROTEIN |
 |
Organism: Human alphaherpesvirus 3
Method: ELECTRON MICROSCOPY Release Date: 2025-04-30 Classification: VIRUS |
 |
Organism: Human alphaherpesvirus 3
Method: ELECTRON MICROSCOPY Resolution:3.70 Å Release Date: 2025-04-30 Classification: VIRAL PROTEIN |
 |
Organism: Human alphaherpesvirus 3
Method: ELECTRON MICROSCOPY Resolution:4.80 Å Release Date: 2025-04-30 Classification: VIRAL PROTEIN |
 |
Organism: Human alphaherpesvirus 3
Method: ELECTRON MICROSCOPY Release Date: 2025-04-30 Classification: VIRUS |
 |
Cryo-Em Structure Of Saccharomyces Cerevisiae Bc1 Complex In Pyraclostrobin-Bound State
Organism: Saccharomyces cerevisiae
Method: ELECTRON MICROSCOPY Release Date: 2024-12-25 Classification: MEMBRANE PROTEIN Ligands: 6PH, 9PE, HEM, 8PE, CN5, UQ6, A1D6K, 7PH, CN3 |
 |
Cryo-Em Structure Of Saccharomyces Cerevisiae Bc1 Complex In Yf23694-Bound State
Organism: Saccharomyces cerevisiae
Method: ELECTRON MICROSCOPY Release Date: 2024-12-25 Classification: MEMBRANE PROTEIN Ligands: 6PH, CN3, 9PE, HEM, CN5, UQ6, A1D6O, 8PE |
 |
Cryo-Em Structure Of Saccharomyces Cerevisiae Bc1 Complex In Metyltetraprole-Bound State
Organism: Saccharomyces cerevisiae
Method: ELECTRON MICROSCOPY Release Date: 2024-12-25 Classification: MEMBRANE PROTEIN Ligands: A1D6P, CN3, 9PE, HEM, 8PE, UQ6, 6PH, FES, CN5 |
 |
Organism: Sus scrofa
Method: ELECTRON MICROSCOPY Release Date: 2024-12-25 Classification: PROTEIN BINDING Ligands: A1D6K, HEM, PEE, HEC, FES, CDL |
 |
Organism: Sus scrofa
Method: ELECTRON MICROSCOPY Release Date: 2024-12-25 Classification: PROTEIN BINDING Ligands: A1D6P, HEM, PEE, CDL, HEC, FES |

