Search Count: 12
 |
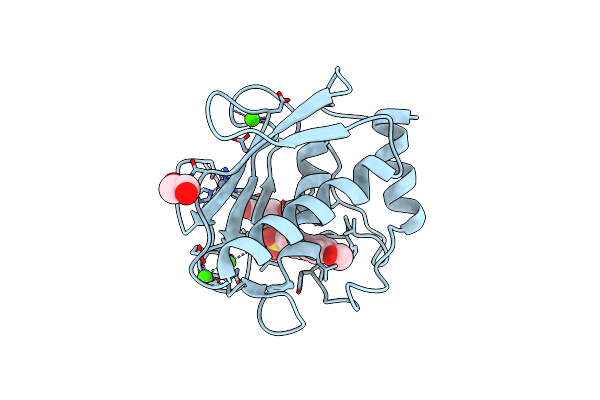
Crystal Structure Of A Dimeric Based Inhibitor Jg34 In Complex With The Mmp-12 Catalytic Domain
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.24 Å Release Date: 2021-10-27 Classification: HYDROLASE/HYDROLASE INHIBITOR Ligands: ZN, CA, 2IF, EDO, PGR |
 |
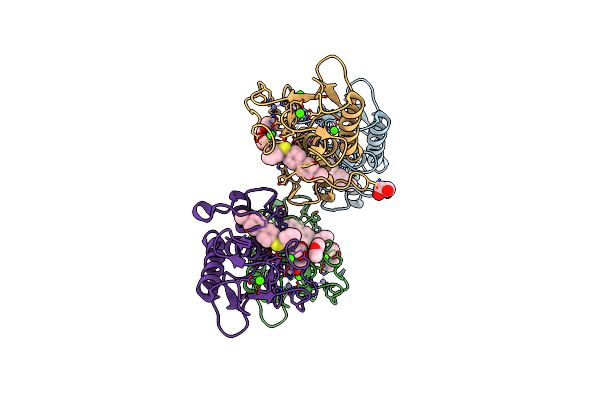
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.20 Å Release Date: 2018-05-16 Classification: HYDROLASE Ligands: ZN, CA, B9N, EDO |
 |
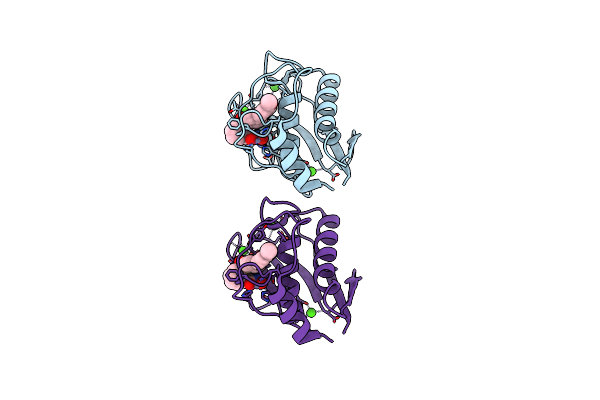
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.49 Å Release Date: 2018-05-16 Classification: HYDROLASE Ligands: ZN, CA, B9Z, DIO, EDO |
 |
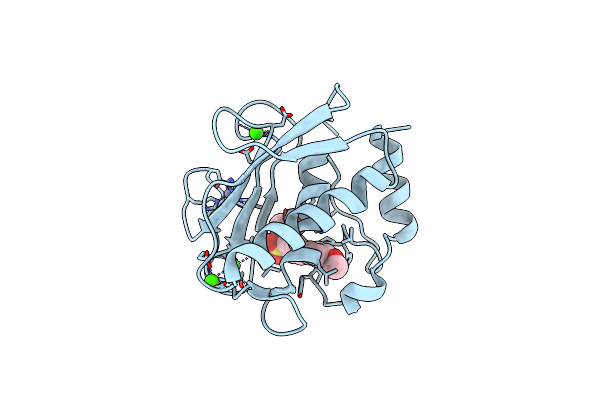
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.59 Å Release Date: 2018-05-16 Classification: HYDROLASE Ligands: LPW, ZN, CA |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.30 Å Release Date: 2018-05-16 Classification: HYDROLASE Ligands: ZN, CA, BKW |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.10 Å Release Date: 2018-05-16 Classification: HYDROLASE Ligands: ZN, CA, B9Z, PZE |
 |
Crystal Structure Of The Catalytic Domain Of Mmp-12 In Complex With A Selective Sugar-Conjugated Arylsulfonamide Carboxylate Water-Soluble Inhibitor (Dc27).
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.45 Å Release Date: 2016-07-06 Classification: HYDROLASE Ligands: H27, ZN, CA, PGO, EDO, PEG, DMS |
 |
Crystal Structure Of The Catalytic Domain Of Mmp-9 In Complex With A Selective Sugar-Conjugated Arylsulfonamide Carboxylate Water-Soluble Inhibitor (Dc27).
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.59 Å Release Date: 2016-07-06 Classification: HYDROLASE Ligands: ZN, CA, H27, EDO, DMS, GOL |
 |
Crystal Structure Of The Catalytic Domain Of Mmp-12 In Complex With A Selective Sugar-Conjugated Triazole-Linked Carboxylate Chelating Water-Soluble Inhibitor (Dc24).
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2016-07-06 Classification: HYDROLASE Ligands: V24, ZN, CA, PGO, EDO, DMS |
 |
Crystal Structure Of The Catalytic Domain Of Mmp-12 In Complex With A Selective Sugar-Conjugated Thiourea-Linked Carboxylate Zinc-Chelator Water-Soluble Inhibitor (Dc31).
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.17 Å Release Date: 2016-07-06 Classification: HYDROLASE Ligands: 67F, ZN, CA, DMS, PGO, DIO, EDO |
 |
Crystal Structure Of The Catalytic Domain Of Mmp-12 In Complex With A Selective Sugar-Conjugated Triazole-Linked Carboxylate Chelator Water-Soluble Inhibitor (Dc32).
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.95 Å Release Date: 2016-07-06 Classification: HYDROLASE Ligands: ZN, CA, 67M, EDO, DMS, PGO |
 |
Crystal Structure Of The Catalytic Domain Of Mmp-12 In Complex With A Selective Sugar-Conjugated Triazole-Linked Carboxylate Zinc-Chelator Water-Soluble Inhibitor (Dc28).
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.05 Å Release Date: 2016-07-06 Classification: HYDROLASE Ligands: V28, ZN, CA |

