Search Count: 8
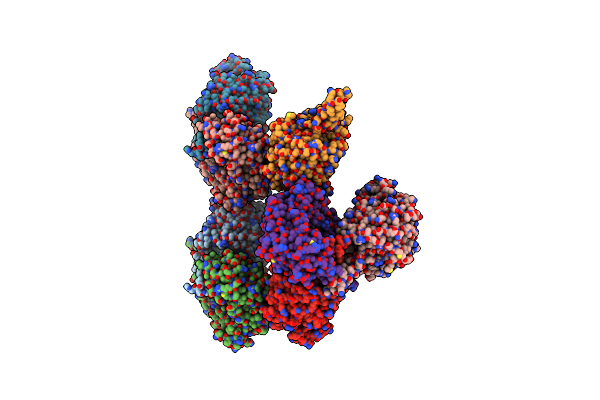 |
P. Syringae Alda Indole-3-Acetaldehyde Dehydrogenase C302A Mutant In Complex With Nad+ And Iaa
Organism: Pseudomonas syringae pv. tomato (strain atcc baa-871 / dc3000)
Method: X-RAY DIFFRACTION Resolution:2.85 Å Release Date: 2021-06-23 Classification: OXIDOREDUCTASE Ligands: NAI, IAC |
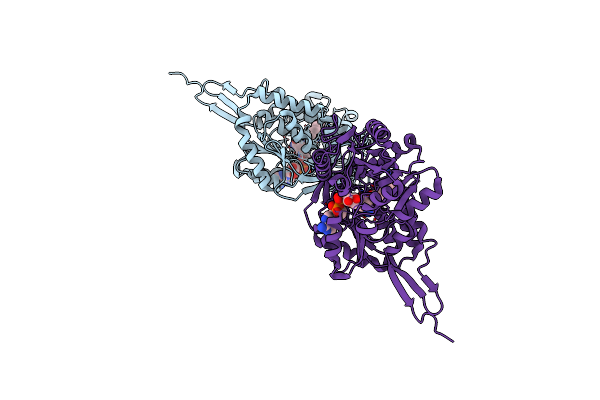 |
Crystal Structure Of Aldehyde Dehydrogenase C (Aldc) Mutant (C291A) From Pseudomonas Syringae In Complexed With Nad+ And Octanal
Organism: Pseudomonas syringae pv. tomato (strain atcc baa-871 / dc3000)
Method: X-RAY DIFFRACTION Resolution:2.52 Å Release Date: 2020-09-09 Classification: OXIDOREDUCTASE Ligands: OYA, NAD |
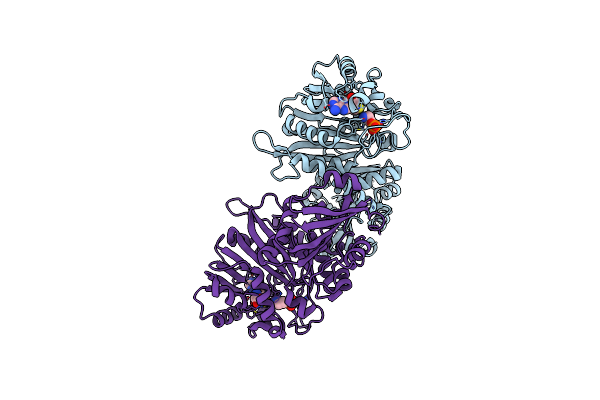 |
Phosphoethanolamine Methyltransferase From The Pine Wilt Nematode Bursaphelenchus Xylophilus
Organism: Bursaphelenchus xylophilus
Method: X-RAY DIFFRACTION Resolution:2.05 Å Release Date: 2020-06-17 Classification: TRANSFERASE Ligands: OPE, SAH |
 |
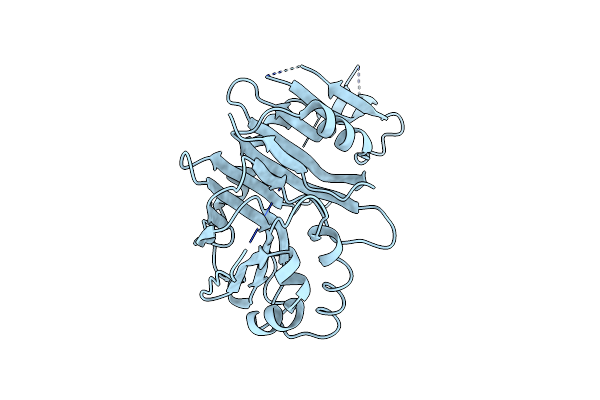
Crystal Structure Of Peruvianin-I (Cysteine Peptidase From Thevetia Peruviana Latex)
Organism: Thevetia peruviana
Method: X-RAY DIFFRACTION Resolution:2.15 Å Release Date: 2020-05-06 Classification: HYDROLASE |
 |
The 3.1 Angstrom Resolution Crystal Structure Of A Mutated Baculovirus P35 After Caspase Cleavage
Organism: Autographa californica nucleopolyhedrovirus
Method: X-RAY DIFFRACTION Resolution:3.10 Å Release Date: 2001-10-24 Classification: APOPTOSIS |
 |
The 2.7 Angstrom Resolution Crystal Structure Of A Mutated Baculovirus P35 After Caspase Cleavage
Organism: Autographa californica nucleopolyhedrovirus
Method: X-RAY DIFFRACTION Resolution:2.70 Å Release Date: 2001-10-24 Classification: APOPTOSIS Ligands: DTT |
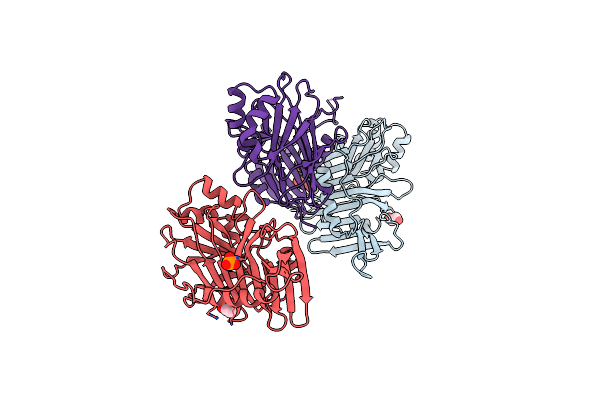 |
Organism: Autographa californica nucleopolyhedrovirus
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 1999-06-01 Classification: APOPTOSIS Ligands: PO4, EDO |
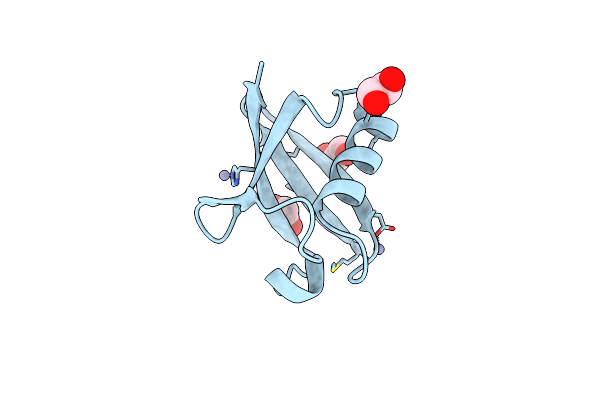 |
Organism: Arabidopsis thaliana
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 1998-12-30 Classification: SIGNALING PROTEIN Ligands: ZN, EDO |

