Search Count: 16
 |
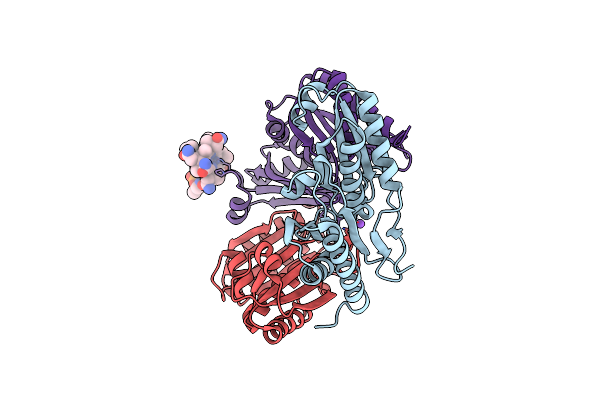
Structure Of Human Volume Regulated Anion Channel Composed Of Swell1 (Lrrc8A)
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2018-08-15 Classification: MEMBRANE PROTEIN |
 |
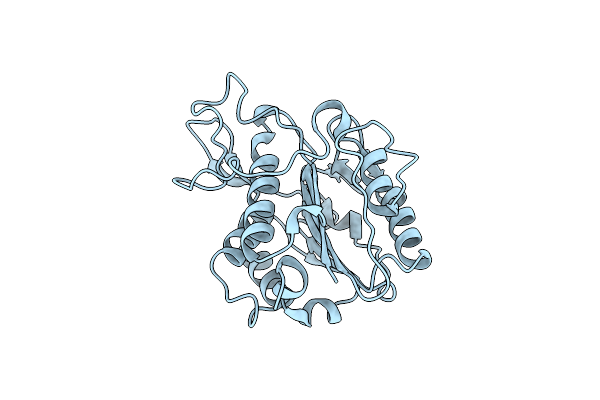
Crystal Structure Of The Eutl Microcompartment Shell Protein From Clostridium Perfringens Bound To Vitamin B12
Organism: Clostridium perfringens
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2014-10-22 Classification: TRANSPORT PROTEIN |
 |
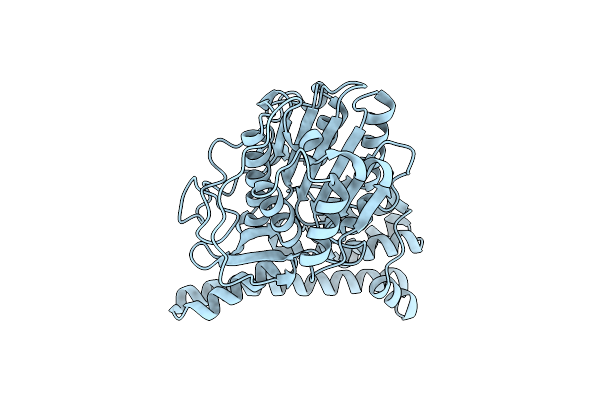
Crystal Structure Of The Mature Form Of Asparaginyl Endopeptidase (Aep)/Legumain Activated At Ph 3.5
Organism: Mus musculus
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2014-02-19 Classification: HYDROLASE |
 |
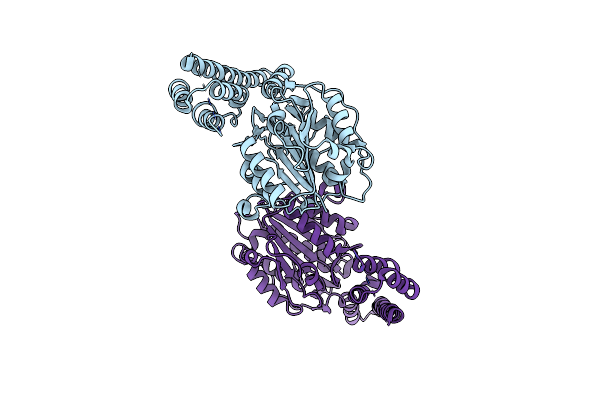
Crystal Structure Of Proenzyme Asparaginyl Endopeptidase (Aep)/Legumain At Ph 7.5
Organism: Mus musculus
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2014-02-19 Classification: HYDROLASE |
 |
Crystal Structure Of Proenzyme Asparaginyl Endopeptidase (Aep)/Legumain Mutant D233A At Ph 7.5
Organism: Mus musculus
Method: X-RAY DIFFRACTION Resolution:2.70 Å Release Date: 2014-02-19 Classification: HYDROLASE |
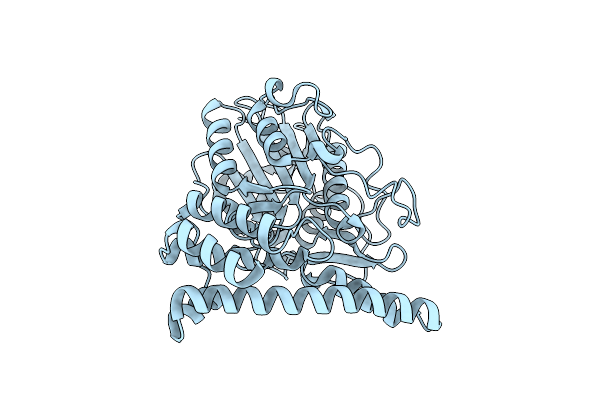 |
Crystal Structure Of Asparaginyl Endopeptidase (Aep)/Legumain Activated At Ph 4.5
Organism: Mus musculus
Method: X-RAY DIFFRACTION Resolution:2.01 Å Release Date: 2014-02-19 Classification: HYDROLASE |
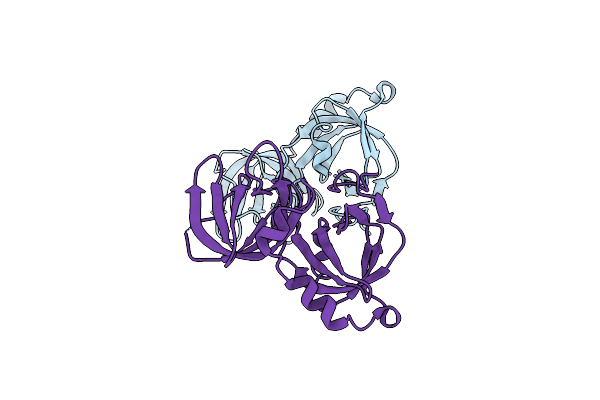 |
Organism: Mus musculus
Method: X-RAY DIFFRACTION Resolution:2.69 Å Release Date: 2013-06-26 Classification: DNA BINDING PROTEIN |
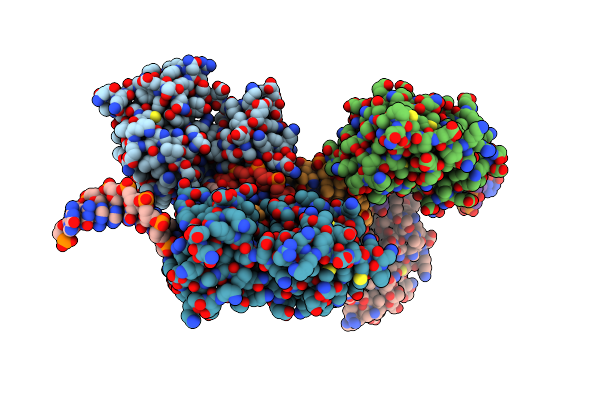 |
Organism: Mus musculus
Method: X-RAY DIFFRACTION Resolution:2.96 Å Release Date: 2013-06-26 Classification: DNA BINDING PROTEIN/DNA |
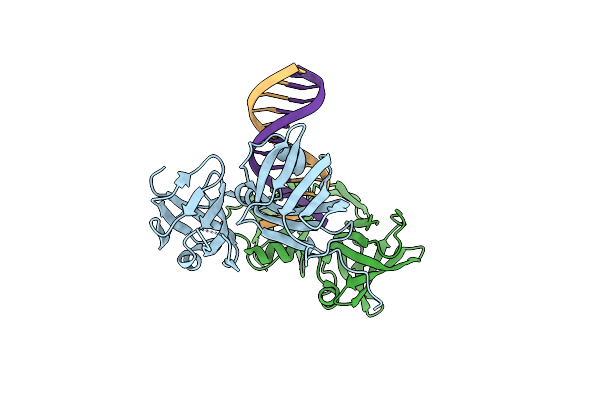 |
Organism: Mus musculus
Method: X-RAY DIFFRACTION Resolution:2.22 Å Release Date: 2013-06-26 Classification: DNA BINDING PROTEIN/DNA |
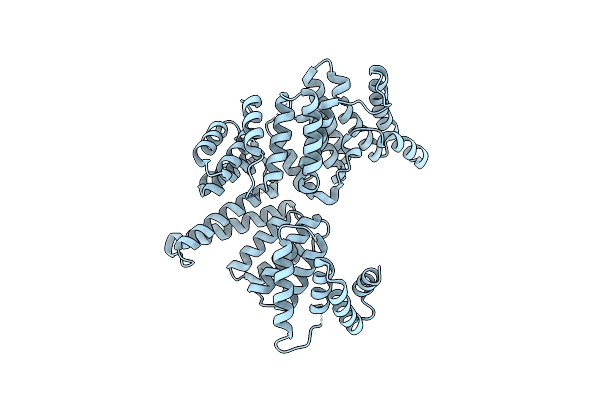 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.97 Å Release Date: 2013-02-13 Classification: RNA BINDING PROTEIN |
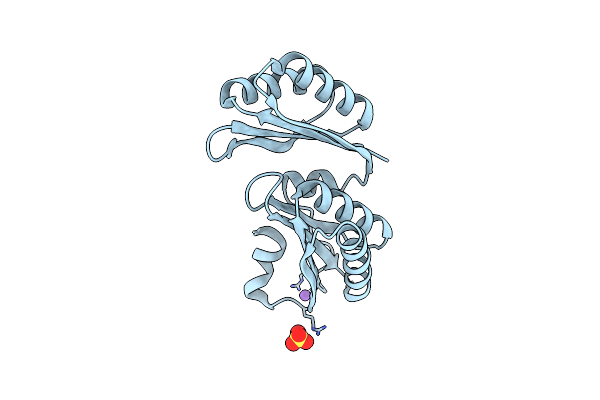 |
Organism: Salmonella enterica subsp. enterica serovar typhimurium
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2010-10-06 Classification: ELECTRON TRANSPORT Ligands: SO4, CL, NA |
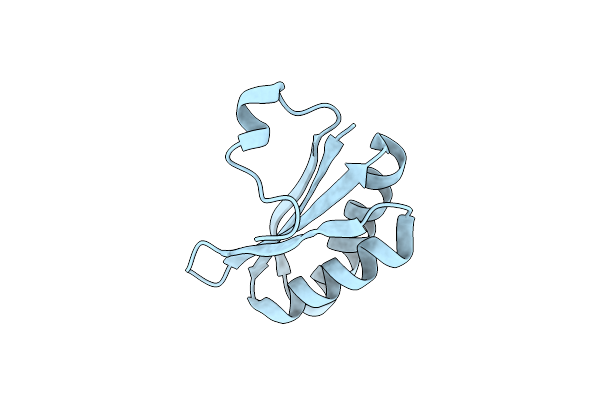 |
Organism: Salmonella enterica subsp. enterica serovar typhimurium
Method: X-RAY DIFFRACTION Resolution:2.26 Å Release Date: 2010-10-06 Classification: UNKNOWN FUNCTION |
 |
Organism: Salmonella typhimurium
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2008-09-02 Classification: UNKNOWN FUNCTION |
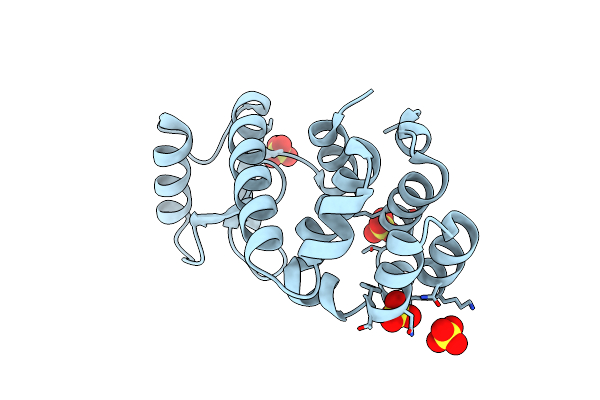 |
Organism: Enterobacteria phage t4
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2006-10-17 Classification: HYDROLASE Ligands: SO4 |
 |
Organism: Enterobacteria phage t4
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2006-10-17 Classification: HYDROLASE Ligands: SO4, GOL |
 |
Organism: Enterobacteria phage t4
Method: X-RAY DIFFRACTION Resolution:2.35 Å Release Date: 2006-10-17 Classification: HYDROLASE Ligands: TRS |

