Search Count: 31
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-12-17 Classification: HYDROLASE |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-12-17 Classification: HYDROLASE |
 |
Organism: Homo sapiens, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2025-12-17 Classification: HYDROLASE Ligands: MN |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2024-12-25 Classification: TRANSCRIPTION Ligands: A1H46 |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2024-12-25 Classification: TRANSCRIPTION Ligands: YNK |
 |
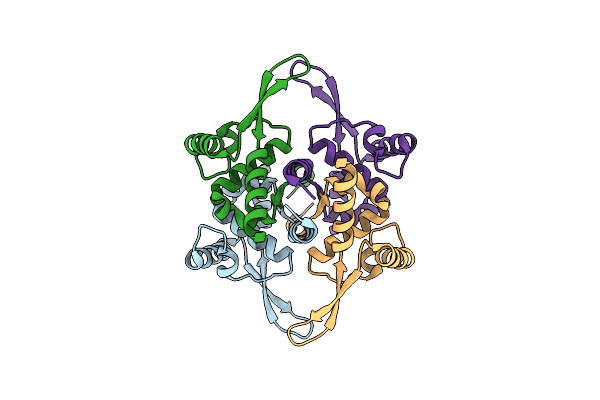
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.59 Å Release Date: 2023-07-19 Classification: MEMBRANE PROTEIN Ligands: NAG |
 |
Organism: Bacillus subtilis subsp. subtilis str. 168
Method: ELECTRON MICROSCOPY Release Date: 2023-05-17 Classification: DNA BINDING PROTEIN |
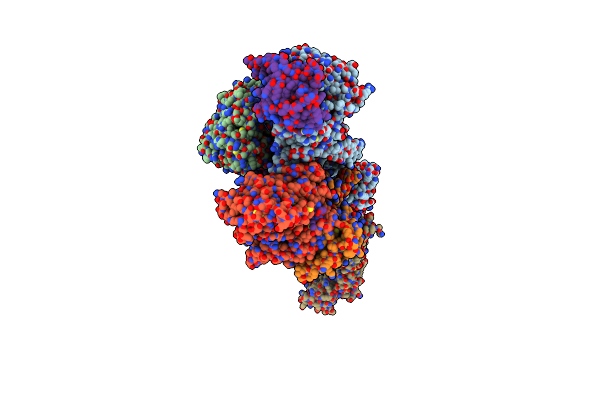 |
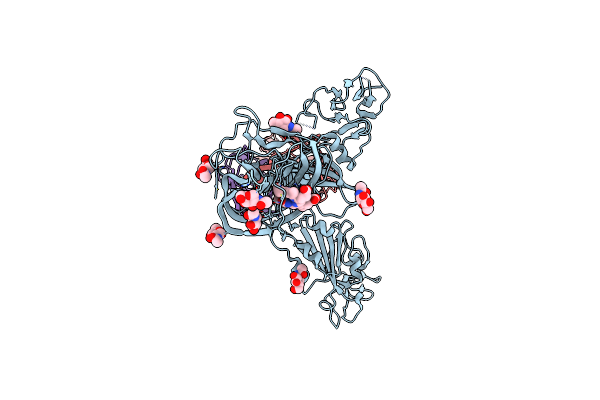
Organism: Saccharomyces cerevisiae (strain atcc 204508 / s288c)
Method: ELECTRON MICROSCOPY Release Date: 2022-09-21 Classification: RECOMBINATION Ligands: ZN |
 |
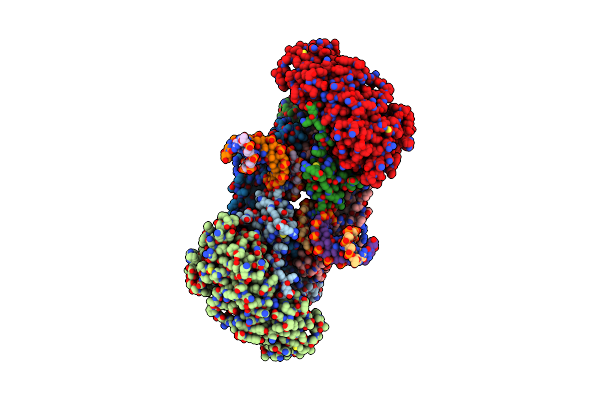
Cryoem Structure Of Sars-Cov-2 Spike Monomer In Complex With Neutralising Antibody P008_60
Organism: Severe acute respiratory syndrome coronavirus 2, Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2022-08-17 Classification: VIRAL PROTEIN Ligands: NAG, 3Q9 |
 |
Structure Of The Stlv Intasome:B56 Complex Bound To The Strand-Transfer Inhibitor Xz450
Organism: Simian t-lymphotropic virus 1, Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2021-08-18 Classification: VIRAL PROTEIN Ligands: ZN, MG, 1L0 |
 |
Stlv-1 Intasome:B56 In Complex With The Strand-Transfer Inhibitor Raltegravir
Organism: Simian t-lymphotropic virus 1, Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2021-08-18 Classification: VIRAL PROTEIN Ligands: ZN, MG, RLT |
 |
Structure Of The Stlv Intasome:B56 Complex Bound To The Strand-Transfer Inhibitor Bictegravir
Organism: Simian t-lymphotropic virus 1, Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2021-08-18 Classification: VIRAL PROTEIN Ligands: ZN, MG, KLQ |
 |
Structural Basis Of Cullin-2 Ring E3 Ligase Regulation By The Cop9 Signalosome
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2019-08-28 Classification: LIGASE Ligands: ZN |
 |
Structural Basis Of Cullin-2 Ring E3 Ligase Regulation By The Cop9 Signalosome
|
 |
Structural Basis Of Cullin-2 Ring E3 Ligase Regulation By The Cop9 Signalosome
|
 |
Structural Basis Of Cullin-2 Ring E3 Ligase Regulation By The Cop9 Signalosome
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2019-08-28 Classification: LIGASE Ligands: ZN |
 |
Structural Basis Of Cullin-2 Ring E3 Ligase Regulation By The Cop9 Signalosome
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2019-08-28 Classification: LIGASE Ligands: ZN |
 |
Atomic-Resolution Structures Of The Apc/C Subunits Apc4 And The Apc5 N-Terminal Domain
Organism: Xenopus laevis
Method: X-RAY DIFFRACTION Resolution:3.20 Å Release Date: 2015-09-02 Classification: CELL CYCLE |
 |
Atomic-Resolution Structures Of The Apc/C Subunits Apc4 And The Apc5 N-Terminal Domain
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:3.40 Å Release Date: 2015-09-02 Classification: CELL CYCLE |
 |
Atomic-Resolution Structures Of The Apc/C Subunits Apc4 And The Apc5 N-Terminal Domain
Organism: Xenopus laevis
Method: X-RAY DIFFRACTION Resolution:2.18 Å Release Date: 2015-09-02 Classification: CELL CYCLE Ligands: EDO |

