Search Count: 168
 |
Organism: Borreliella burgdorferi
Method: X-RAY DIFFRACTION Release Date: 2025-03-12 Classification: OXIDOREDUCTASE Ligands: 6V0, OXM, CA, FBP |
 |
High-Resolution Structure Of Cytochrome C Peroxidase From Yeast At Ambient Temperature And Ambient Pressure
Organism: Saccharomyces cerevisiae (strain atcc 204508 / s288c)
Method: X-RAY DIFFRACTION Resolution:1.54 Å Release Date: 2024-10-16 Classification: OXIDOREDUCTASE Ligands: HEM |
 |
High-Resolution Structure Of Cytochrome C Peroxidase From Yeast Under Cryogenic Conditions And Ambient Pressure
Organism: Saccharomyces cerevisiae (strain atcc 204508 / s288c)
Method: X-RAY DIFFRACTION Resolution:1.78 Å Release Date: 2024-10-16 Classification: OXIDOREDUCTASE Ligands: HEM |
 |
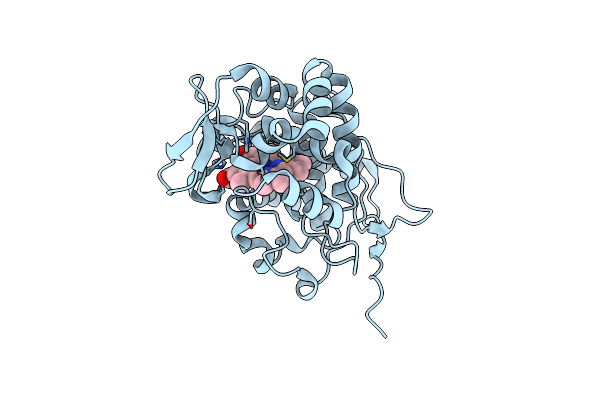
High-Resolution Structure Of Cytochrome C Peroxidase From Yeast At Ambient Temperature And 1.5 Kbar
Organism: Saccharomyces cerevisiae s288c
Method: X-RAY DIFFRACTION Resolution:2.31 Å Release Date: 2024-10-16 Classification: OXIDOREDUCTASE Ligands: HEM, PER |
 |
High-Resolution Structure Of Cytochrome C Peroxidase From Yeast At Ambient Temperature And 3.0 Kbar
Organism: Saccharomyces cerevisiae (strain atcc 204508 / s288c)
Method: X-RAY DIFFRACTION Resolution:2.31 Å Release Date: 2024-10-16 Classification: OXIDOREDUCTASE Ligands: HEM |
 |
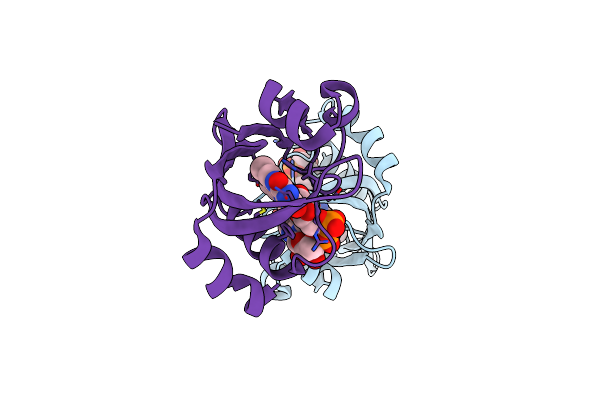
Organism: Homo sapiens, Drosophila melanogaster
Method: ELECTRON MICROSCOPY Release Date: 2023-02-15 Classification: CIRCADIAN CLOCK PROTEIN Ligands: FAD |
 |
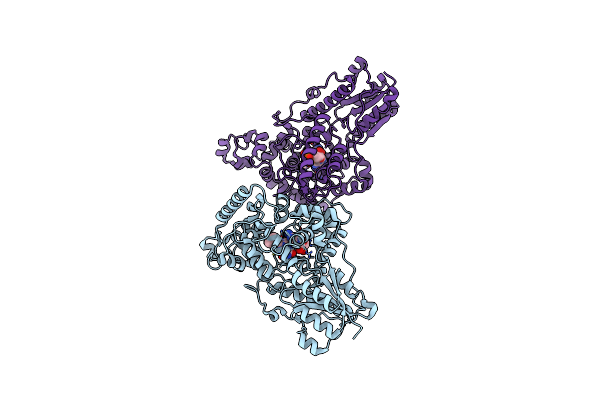
Redox Properties And Pas Domain Structure Of The E. Coli Energy Sensor Aer Indicate A Multi-State Sensing Mechanism
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2022-11-09 Classification: SIGNALING PROTEIN Ligands: FAD |
 |
Organism: Drosophila melanogaster
Method: X-RAY DIFFRACTION Resolution:3.50 Å Release Date: 2022-08-24 Classification: CIRCADIAN CLOCK PROTEIN Ligands: FAD |
 |
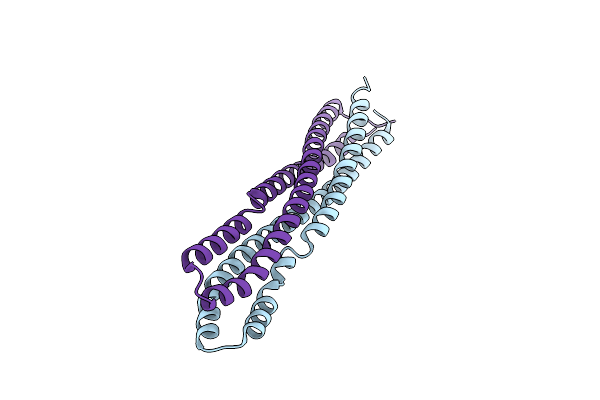
Organism: Drosophila melanogaster
Method: X-RAY DIFFRACTION Resolution:2.58 Å Release Date: 2021-05-05 Classification: CIRCADIAN CLOCK PROTEIN Ligands: MG, FAD |
 |
Organism: Treponema denticola
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2020-10-28 Classification: SIGNALING PROTEIN |
 |
Yeast Cytochrome C Peroxidase (W191Y:L232E) In Complex With Iso-1 Cytochrome C
Organism: Saccharomyces cerevisiae (strain atcc 204508 / s288c)
Method: X-RAY DIFFRACTION Resolution:2.90 Å Release Date: 2019-10-23 Classification: Electron transport/Oxidoreductase Ligands: HEM |
 |
Yeast Cytochrome C Peroxidase (W191Y:L232H) In Complex With Iso-1 Cytochrome C
Organism: Saccharomyces cerevisiae (strain atcc 204508 / s288c)
Method: X-RAY DIFFRACTION Resolution:2.91 Å Release Date: 2019-10-23 Classification: Electron transport/Oxidoreductase Ligands: HEM |
 |
Organism: Saccharomyces cerevisiae (strain atcc 204508 / s288c)
Method: X-RAY DIFFRACTION Resolution:1.91 Å Release Date: 2019-10-23 Classification: Electron transport/Oxidoreductase Ligands: HEM |
 |
Organism: Treponema denticola
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2019-08-21 Classification: MOTOR PROTEIN |
 |
Organism: Treponema denticola
Method: X-RAY DIFFRACTION Resolution:1.42 Å Release Date: 2019-08-14 Classification: MOTOR PROTEIN |
 |
Crystal Structure Of The Wild Type D2 Domain (A168-T344) Of The Flagellar Hook Protein Flge From Treponema Denticola
Organism: Treponema denticola
Method: X-RAY DIFFRACTION Resolution:1.73 Å Release Date: 2019-08-14 Classification: MOTOR PROTEIN |
 |
Organism: Treponema denticola
Method: X-RAY DIFFRACTION Resolution:3.04 Å Release Date: 2019-08-14 Classification: MOTOR PROTEIN |
 |
Organism: Treponema denticola atcc 35404
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2019-06-19 Classification: SIGNALING PROTEIN |
 |
Organism: Thermotoga maritima
Method: X-RAY DIFFRACTION Resolution:2.56 Å Release Date: 2019-06-19 Classification: SIGNALING PROTEIN |
 |
Organism: Thermotoga maritima
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2019-06-19 Classification: SIGNALING PROTEIN Ligands: ZN |

