Search Count: 16
 |
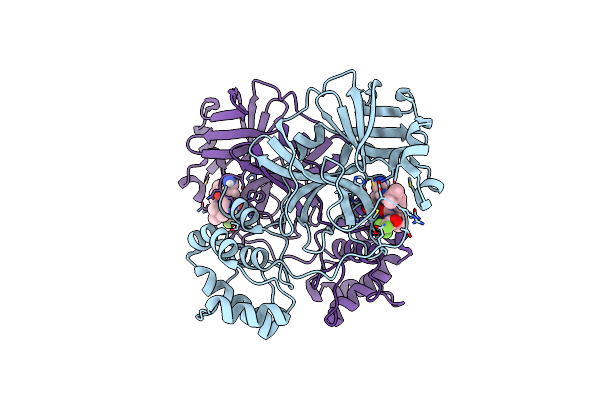
Structure Of Sars-Cov-2 Mpro Mutant (S144A) In Complex With Nirmatrelvir (Pf-07321332)
Organism: Severe acute respiratory syndrome coronavirus 2
Method: X-RAY DIFFRACTION Resolution:1.49 Å Release Date: 2024-08-07 Classification: VIRAL PROTEIN, HYDROLASE/INHIBITOR Ligands: 4WI |
 |
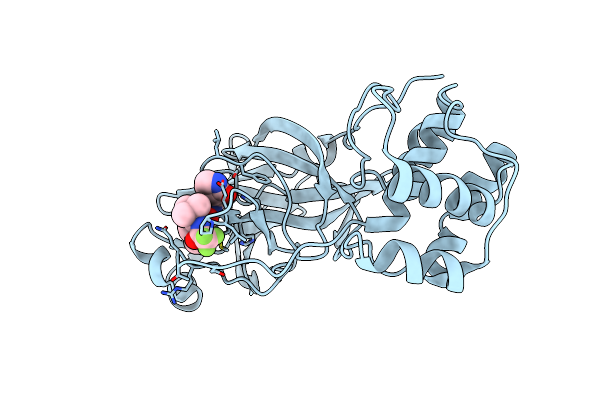
Structure Of Sars-Cov-2 Mpro Mutant (A173V) In Complex With Nirmatrelvir (Pf-07321332)
Organism: Severe acute respiratory syndrome coronavirus 2
Method: X-RAY DIFFRACTION Resolution:1.88 Å Release Date: 2024-08-07 Classification: VIRAL PROTEIN, HYDROLASE/INHIBITOR Ligands: 4WI |
 |
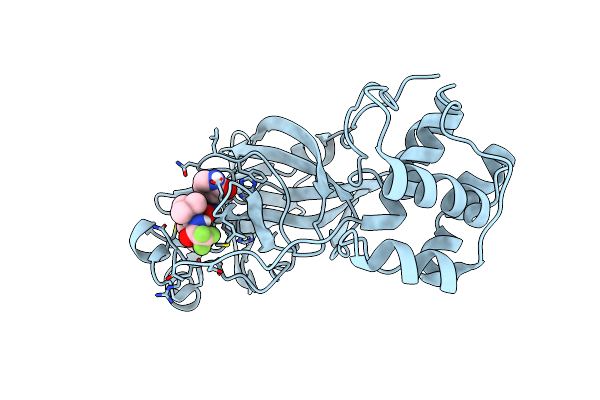
Structure Of Sars-Cov-2 Mpro Mutant (A173V,T304I)) In Complex With Nirmatrelvir (Pf-07321332)
Organism: Severe acute respiratory syndrome coronavirus 2
Method: X-RAY DIFFRACTION Resolution:2.42 Å Release Date: 2024-08-07 Classification: VIRAL PROTEIN, HYDROLASE/INHIBITOR Ligands: 4WI |
 |
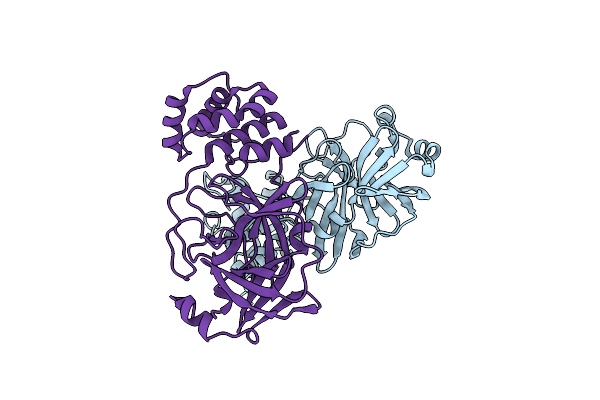
Structure Of Sars-Cov-2 Mpro Mutant (T21I,S144A,T304I) In Complex With Nirmatrelvir (Pf-07321332)
Organism: Severe acute respiratory syndrome coronavirus 2
Method: X-RAY DIFFRACTION Resolution:1.54 Å Release Date: 2024-08-07 Classification: VIRAL PROTEIN, HYDROLASE/INHIBITOR Ligands: 4WI |
 |
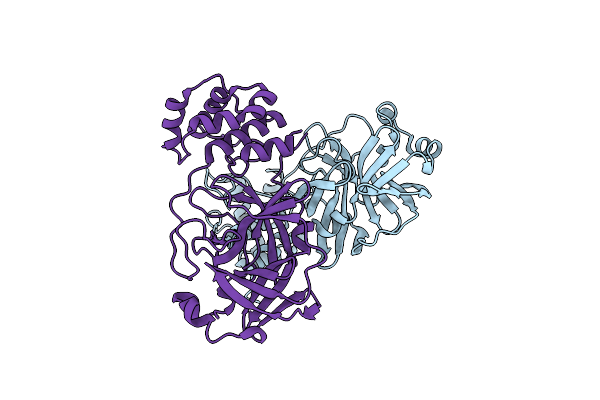
Organism: Severe acute respiratory syndrome coronavirus 2
Method: X-RAY DIFFRACTION Resolution:2.29 Å Release Date: 2024-08-07 Classification: VIRAL PROTEIN, HYDROLASE |
 |
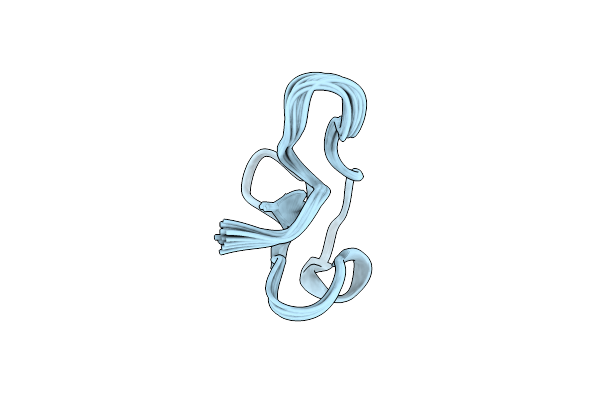
Organism: Severe acute respiratory syndrome coronavirus 2
Method: X-RAY DIFFRACTION Resolution:2.42 Å Release Date: 2024-08-07 Classification: VIRAL PROTEIN, HYDROLASE Ligands: HOH |
 |
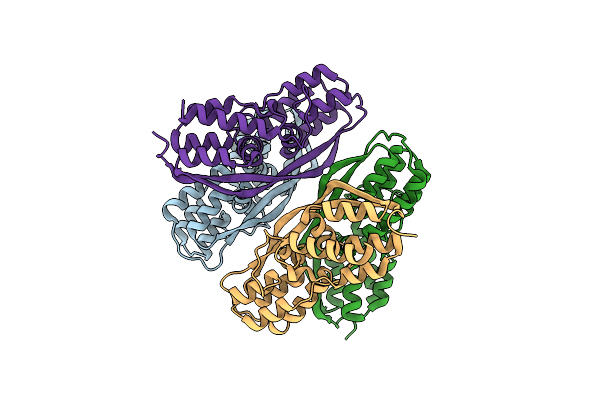
Organism: Synthetic construct
Method: SOLUTION NMR Release Date: 2014-05-07 Classification: PROTEIN BINDING |
 |
Organism: Thermoproteus tenax
Method: X-RAY DIFFRACTION Resolution:2.60 Å Release Date: 2014-03-19 Classification: TRANSCRIPTION |
 |
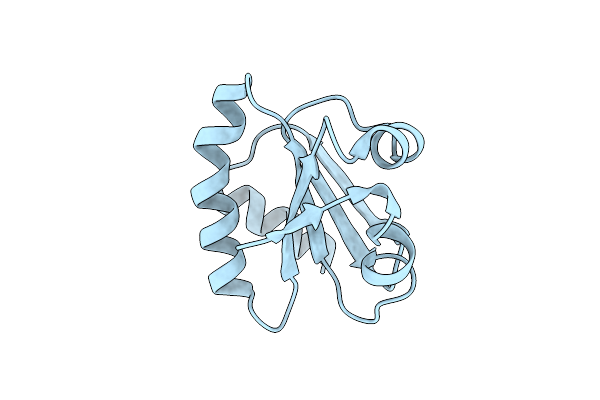
Crystal Structure Of A Pentameric Capsid Protein Isolated From Metagenomic Phage Sequences Solved By Iodide Sad Phasing
Organism: Unidentified
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2013-03-27 Classification: VIRAL PROTEIN Ligands: IOD, NA |
 |
Organism: Silicibacter phage dss3phi2
Method: X-RAY DIFFRACTION Resolution:1.75 Å Release Date: 2013-01-23 Classification: ELECTRON TRANSPORT |
 |
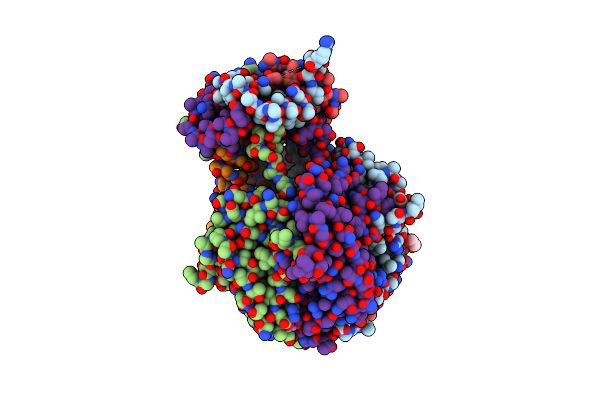
Iodide Sad Phased Crystal Structure Of A Phosphoglucomutase From Brucella Melitensis Complexed With Glucose-6-Phosphate
Organism: Brucella melitensis bv. 1
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2012-11-21 Classification: ISOMERASE Ligands: IOD, G6Q, EDO, MG |
 |
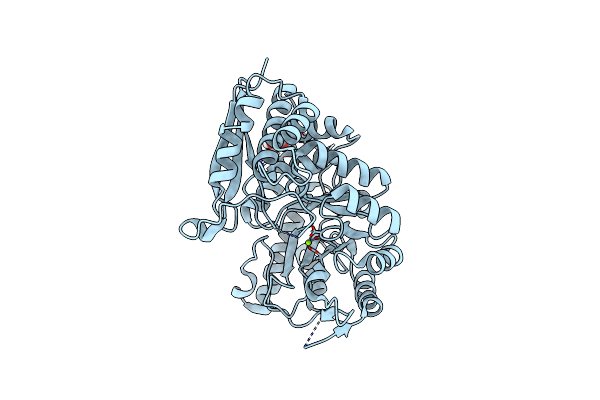
Crystal Structure Of A Pentameric Capsid Protein Isolated From Metagenomic Phage Sequences (Casp)
Organism: Unidentified
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2012-08-29 Classification: VIRAL PROTEIN Ligands: EDO, NA |
 |
Organism: Trypanosoma cruzi
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2012-08-29 Classification: LYASE Ligands: MG, EDO |
 |
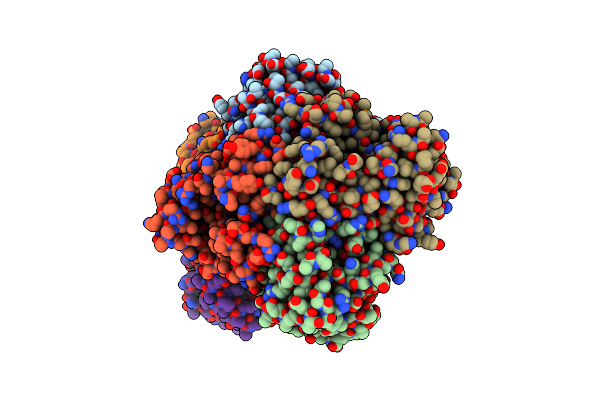
Crystal Structure Of A Hypothetical Metallo-Beta-Lactamase From Burkholderia Pseudomallei
Organism: Burkholderia pseudomallei
Method: X-RAY DIFFRACTION Release Date: 2012-05-09 Classification: HYDROLASE Ligands: CA, CL, EDO, GOL, SO4 |
 |
Crystal Structure Of An Enoyl-(Acyl Carrier Protein) Reductase From Bartonella Henselae
Organism: Bartonella henselae
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2012-04-18 Classification: OXIDOREDUCTASE |
 |
Organism: Mus musculus
Method: X-RAY DIFFRACTION Resolution:1.44 Å Release Date: 2010-09-29 Classification: PROTEIN TRANSPORT Ligands: CA |

