Search Count: 27
 |
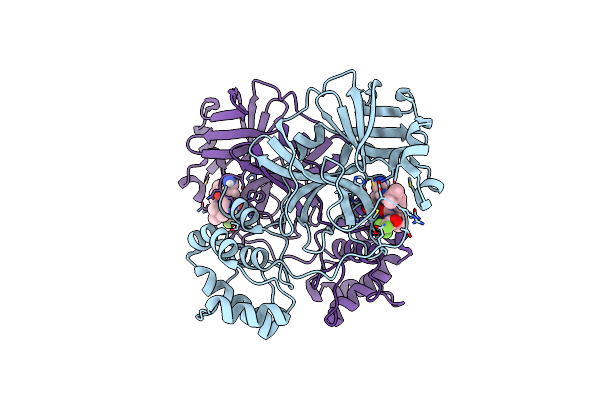
Structure Of Sars-Cov-2 Mpro Mutant (S144A) In Complex With Nirmatrelvir (Pf-07321332)
Organism: Severe acute respiratory syndrome coronavirus 2
Method: X-RAY DIFFRACTION Resolution:1.49 Å Release Date: 2024-08-07 Classification: VIRAL PROTEIN, HYDROLASE/INHIBITOR Ligands: 4WI |
 |
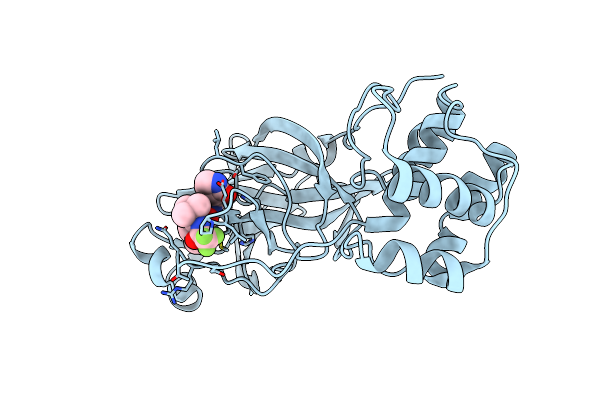
Structure Of Sars-Cov-2 Mpro Mutant (A173V) In Complex With Nirmatrelvir (Pf-07321332)
Organism: Severe acute respiratory syndrome coronavirus 2
Method: X-RAY DIFFRACTION Resolution:1.88 Å Release Date: 2024-08-07 Classification: VIRAL PROTEIN, HYDROLASE/INHIBITOR Ligands: 4WI |
 |
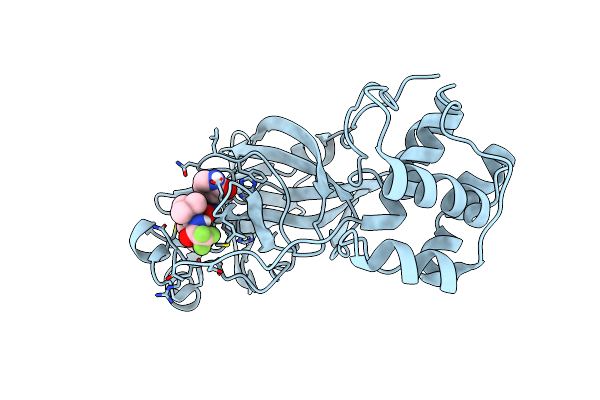
Structure Of Sars-Cov-2 Mpro Mutant (A173V,T304I)) In Complex With Nirmatrelvir (Pf-07321332)
Organism: Severe acute respiratory syndrome coronavirus 2
Method: X-RAY DIFFRACTION Resolution:2.42 Å Release Date: 2024-08-07 Classification: VIRAL PROTEIN, HYDROLASE/INHIBITOR Ligands: 4WI |
 |
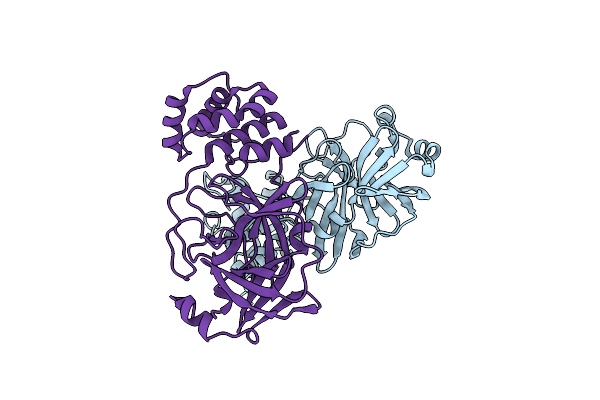
Structure Of Sars-Cov-2 Mpro Mutant (T21I,S144A,T304I) In Complex With Nirmatrelvir (Pf-07321332)
Organism: Severe acute respiratory syndrome coronavirus 2
Method: X-RAY DIFFRACTION Resolution:1.54 Å Release Date: 2024-08-07 Classification: VIRAL PROTEIN, HYDROLASE/INHIBITOR Ligands: 4WI |
 |
Organism: Severe acute respiratory syndrome coronavirus 2
Method: X-RAY DIFFRACTION Resolution:2.29 Å Release Date: 2024-08-07 Classification: VIRAL PROTEIN, HYDROLASE |
 |
Organism: Severe acute respiratory syndrome coronavirus 2
Method: X-RAY DIFFRACTION Resolution:2.42 Å Release Date: 2024-08-07 Classification: VIRAL PROTEIN, HYDROLASE Ligands: HOH |
 |
Structure Of Sars-Cov-2 Mpro Lambda (G15S) In Complex With Nirmatrelvir (Pf-07321332)
Organism: Severe acute respiratory syndrome coronavirus 2
Method: X-RAY DIFFRACTION Resolution:1.68 Å Release Date: 2022-03-09 Classification: HYDROLASE/HYDROLASE INHIBITOR Ligands: 4WI |
 |
Structure Of Sars-Cov-2 Mpro Mutant (K90R) In Complex With Nirmatrelvir (Pf-07321332)
Organism: Severe acute respiratory syndrome coronavirus 2
Method: X-RAY DIFFRACTION Resolution:2.09 Å Release Date: 2022-03-09 Classification: HYDROLASE/HYDROLASE INHIBITOR Ligands: 4WI |
 |
Structure Of Sars-Cov-2 Mpro Omicron P132H In Complex With Nirmatrelvir (Pf-07321332)
Organism: Severe acute respiratory syndrome coronavirus 2
Method: X-RAY DIFFRACTION Resolution:1.63 Å Release Date: 2022-01-26 Classification: HYDROLASE/HYDROLASE INHIBITOR Ligands: 4WI |
 |
Organism: Saccharomyces cerevisiae (strain atcc 204508 / s288c)
Method: ELECTRON MICROSCOPY Release Date: 2021-06-30 Classification: DNA BINDING PROTEIN Ligands: SF4 |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2021-01-13 Classification: PROTEIN BINDING Ligands: SO4 |
 |
Organism: Synthetic construct
Method: SOLUTION NMR Release Date: 2014-05-07 Classification: PROTEIN BINDING |
 |
Organism: Thermoproteus tenax
Method: X-RAY DIFFRACTION Resolution:2.60 Å Release Date: 2014-03-19 Classification: TRANSCRIPTION |
 |
Crystal Structure Of A Pentameric Capsid Protein Isolated From Metagenomic Phage Sequences Solved By Iodide Sad Phasing
Organism: Unidentified
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2013-03-27 Classification: VIRAL PROTEIN Ligands: IOD, NA |
 |
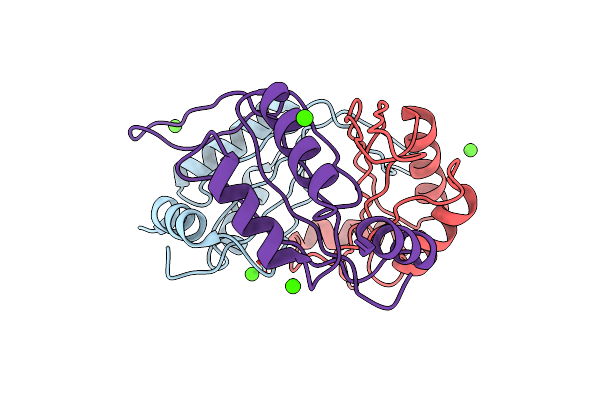
Organism: Synechococcus elongatus
Method: X-RAY DIFFRACTION Resolution:1.55 Å Release Date: 2013-03-06 Classification: HYDROLASE Ligands: ZN, EDO, FMT, CL |
 |
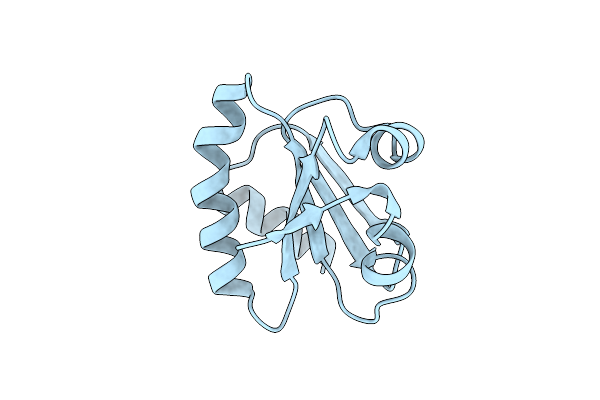
Crystal Structure Of Divalent Ion Tolerance Protein Cuta1 From Ehrlichia Chaffeensis
Organism: Ehrlichia chaffeensis
Method: X-RAY DIFFRACTION Resolution:2.55 Å Release Date: 2013-02-13 Classification: PROTEIN BINDING Ligands: CA |
 |
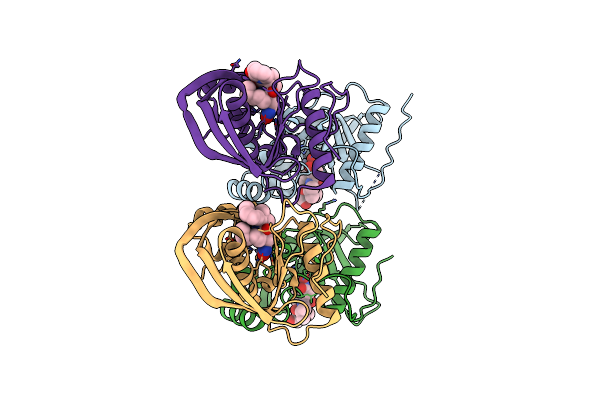
Organism: Silicibacter phage dss3phi2
Method: X-RAY DIFFRACTION Resolution:1.75 Å Release Date: 2013-01-23 Classification: ELECTRON TRANSPORT |
 |
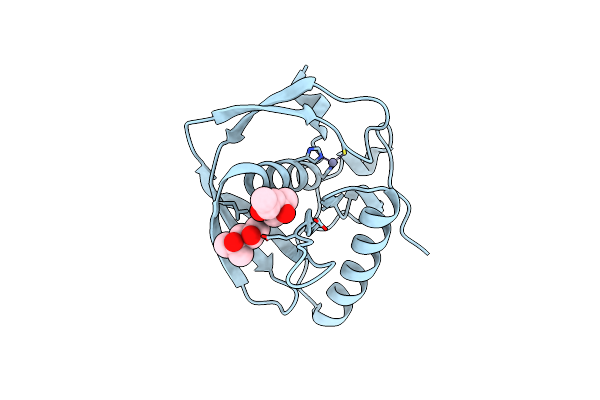
Crystal Structure Of A Peptide Deformylase From Synechococcus Elongatus In Complex With Actinonin
Organism: Synechococcus elongatus
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2013-01-16 Classification: HYDROLASE/HYDROLASE INHIBITOR Ligands: ZN, BR, BB2 |
 |
Crystal Structure Of A Probable Peptide Deformylase From Synechococcus Phage S-Ssm7
Organism: Synechococcus phage s-ssm7
Method: X-RAY DIFFRACTION Resolution:1.95 Å Release Date: 2013-01-09 Classification: HYDROLASE Ligands: ZN, MRD |
 |
Crystal Structure Of A Probable Peptide Deformylase From Strucynechococcus Phage S-Ssm7 In Complex With Actinonin
Organism: Synechococcus phage s-ssm7
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2013-01-09 Classification: HYDROLASE/ANTIBIOTIC Ligands: ZN, CL, EDO, BB2 |

