Search Count: 24
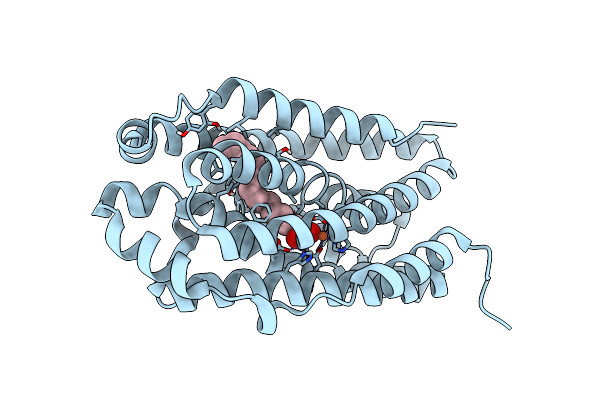 |
R2-Like Ligand-Binding Oxidase G68F Mutant With Aerobically Reconstituted Mn/Fe Cofactor
Organism: Geobacillus kaustophilus (strain hta426)
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2019-10-16 Classification: OXIDOREDUCTASE Ligands: PLM, MN3, MN, FE |
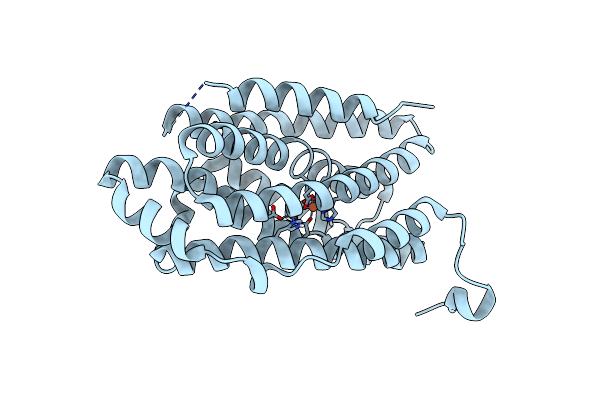 |
R2-Like Ligand-Binding Oxidase G68F Mutant With Anaerobically Reconstituted Mn/Fe Cofactor
Organism: Geobacillus kaustophilus (strain hta426)
Method: X-RAY DIFFRACTION Resolution:1.85 Å Release Date: 2019-10-16 Classification: OXIDOREDUCTASE Ligands: MN, FE2 |
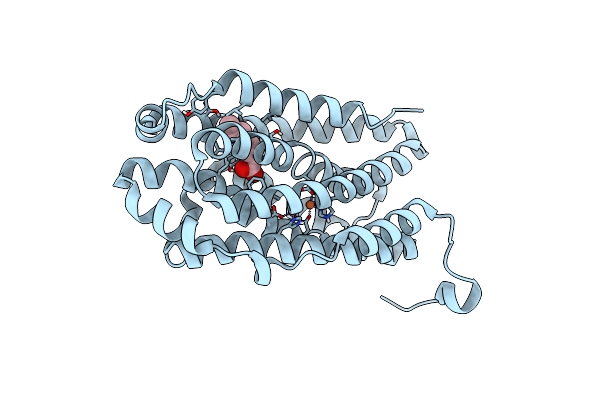 |
R2-Like Ligand-Binding Oxidase G68L Mutant With Aerobically Reconstituted Fe/Fe Cofactor
Organism: Geobacillus kaustophilus (strain hta426)
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2019-10-16 Classification: OXIDOREDUCTASE Ligands: FE, OCA |
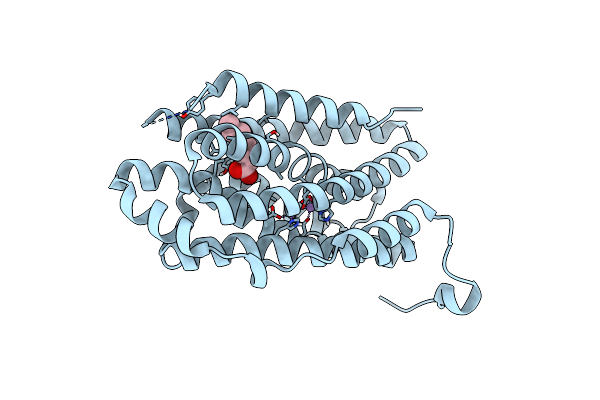 |
R2-Like Ligand-Binding Oxidase G68L Mutant With Non-Activated Mn/Mn Cofactor (After Aerobic Reconstitution With Mn And Fe)
Organism: Geobacillus kaustophilus (strain hta426)
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2019-10-16 Classification: OXIDOREDUCTASE Ligands: MN, OCA |
 |
R2-Like Ligand-Binding Oxidase G68L Mutant With Anaerobically Reconstituted Mn/Fe Cofactor
Organism: Geobacillus kaustophilus (strain hta426)
Method: X-RAY DIFFRACTION Resolution:1.65 Å Release Date: 2019-10-16 Classification: OXIDOREDUCTASE Ligands: MN, FE2, OCA |
 |
Organism: Thermosynechococcus elongatus (strain bp-1)
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2019-01-16 Classification: ELECTRON TRANSPORT Ligands: ACY |
 |
Structure Of The Photosynthetic Complex I From Thermosynechococcus Elongatus
Organism: Thermosynechococcus elongatus bp-1
Method: ELECTRON MICROSCOPY Release Date: 2019-01-09 Classification: PROTON TRANSPORT Ligands: BCR, LMG, SF4 |
 |
Ribonucleotide Reductase Class Ie R2 From Mesoplasma Florum, Dopa-Active Form
Organism: Mesoplasma florum l1
Method: X-RAY DIFFRACTION Release Date: 2018-08-22 Classification: OXIDOREDUCTASE Ligands: CA |
 |
Organism: Mesoplasma florum (strain atcc 33453 / nbrc 100688 / nctc 11704 / l1)
Method: X-RAY DIFFRACTION Resolution:1.23 Å Release Date: 2018-08-22 Classification: OXIDOREDUCTASE Ligands: CA |
 |
Organism: Homo sapiens, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.90 Å Release Date: 2018-05-09 Classification: SIGNALING PROTEIN Ligands: NAG |
 |
Organism: Homo sapiens, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:3.00 Å Release Date: 2018-05-09 Classification: SIGNALING PROTEIN Ligands: NAG |
 |
Organism: Homo sapiens, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:3.35 Å Release Date: 2018-05-09 Classification: PEPTIDE BINDING PROTEIN Ligands: NAG |
 |
Organism: Homo sapiens, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.67 Å Release Date: 2018-05-09 Classification: SIGNALING PROTEIN Ligands: NAG |
 |
Crystal Structure Of The Catalytic Domain Of Membrane Type 1 Matrix Metalloproteinase
Organism: Homo sapiens, Unidentified
Method: X-RAY DIFFRACTION Resolution:2.24 Å Release Date: 2017-10-04 Classification: HYDROLASE Ligands: CA, ZN, EPE, GOL |
 |
Organism: Unidentified, Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.86 Å Release Date: 2016-09-14 Classification: Hydrolase/RNA Ligands: 0G6, NAG, EDO, GOL, CA, MRD, MG |
 |
R2-Like Ligand-Binding Oxidase With Aerobically Reconstituted Mn/Fe Cofactor (Reconstituted In Solution)
Organism: Geobacillus kaustophilus (strain hta426)
Method: X-RAY DIFFRACTION Resolution:2.07 Å Release Date: 2016-04-27 Classification: OXIDOREDUCTASE Ligands: MN3, FE, PLM |
 |
Crystal Structure Of The Hemagglutinin From A H1N1Pdm A/Washington/5/2011 Virus
Organism: Influenza a virus
Method: X-RAY DIFFRACTION Resolution:3.00 Å Release Date: 2014-02-26 Classification: VIRAL PROTEIN Ligands: NAG |
 |
R2-Like Ligand-Binding Oxidase With Aerobically Reconstituted Metal Cofactor
Organism: Geobacillus kaustophilus
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2013-10-16 Classification: OXIDOREDUCTASE Ligands: MN3, FE, MN, NA, PLM |
 |
R2-Like Ligand-Binding Oxidase With Anaerobically Reconstituted Metal Cofactor
Organism: Geobacillus kaustophilus
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2013-10-16 Classification: OXIDOREDUCTASE Ligands: MN, FE2, PLM |
 |
Organism: Geobacillus kaustophilus
Method: X-RAY DIFFRACTION Resolution:2.29 Å Release Date: 2013-10-16 Classification: OXIDOREDUCTASE Ligands: PLM |

