Search Count: 27
 |
Crystal Structure Of Human Histidine Triad Nucleotide-Binding Protein 1 In Complex With 5'-O-[(3-Indolyl)-1-Ethyl]Carbamoyl Ethenoadenosine
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2023-06-14 Classification: HYDROLASE Ligands: X7I |
 |
Crystal Structure Of Human Histidine Triad Nucleotide-Binding Protein 1 In Complex With 5'-O-[(3-Indolyl)-1-Ethyl]Carbamoyl 2-Aminoethenoadenosine
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2023-06-14 Classification: HYDROLASE Ligands: XKB |
 |
Crystal Structure Of Human Histidine Triad Nucleotide-Binding Protein 1 In Complex With 5'-O-[(3-Indolyl)-1-Ethyl]Carbamoyl N2-Methyl-2-Aminoethenoadenosine
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2023-06-14 Classification: HYDROLASE Ligands: XKF |
 |
Crystal Structure Of Human Histidine Triad Nucleotide-Binding Protein 1 In Complex With 5'-O-[N-(3-Indolepropionic Acid)Sulfamoyl] 2-Aminoethenoadenosine
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2023-06-14 Classification: HYDROLASE Ligands: XKK |
 |
Crystal Structure Of Human Histidine Triad Nucleotide-Binding Protein 1 In Complex With 5'-O-[N-(3-Indolepropionic Acid)Sulfamoyl] N2-Methyl-2-Aminoethenoadenosine
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2023-06-14 Classification: HYDROLASE Ligands: XKO |
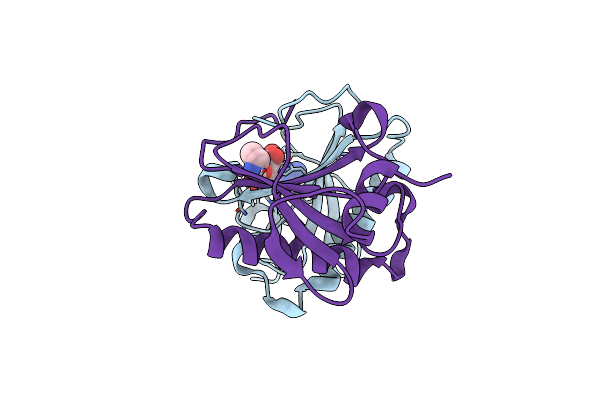 |
Human Histidine Triad Nucleotide Binding Protein 1 (Hint1) With Bound 5'-O-[1-Ethyl]Carbamoyl Guanosine
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.45 Å Release Date: 2019-09-18 Classification: HYDROLASE Ligands: KB7, CL |
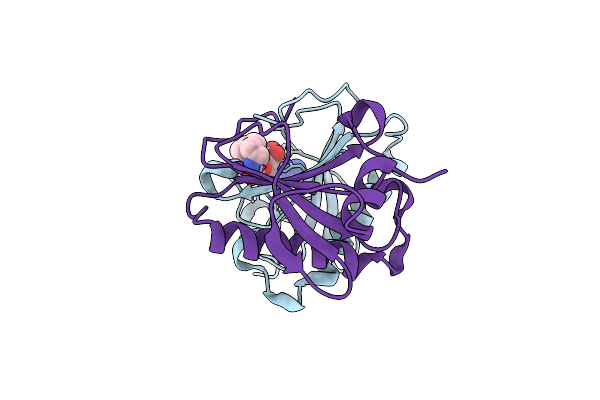 |
Human Histidine Triad Nucleotide Binding Protein 1 (Hint1) With Bound 5'-O-[3-Phenyl-1-Ethyl]Carbamoyl Guanosine
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.75 Å Release Date: 2019-09-18 Classification: HYDROLASE Ligands: KBJ |
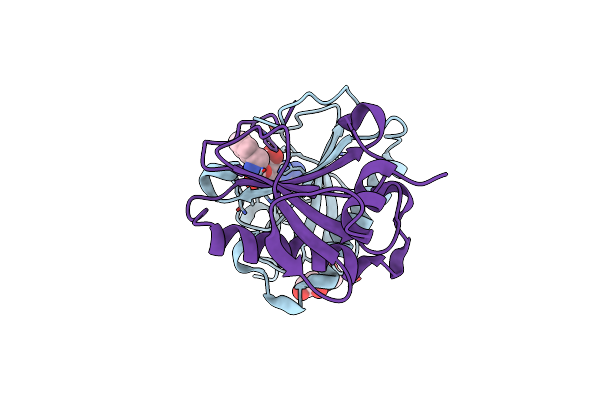 |
Human Histidine Triad Nucleotide Binding Protein 1 (Hint1) With Bound 5'-O-[1-Benzyl]Carbamoyl Guanosine
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.10 Å Release Date: 2019-09-18 Classification: HYDROLASE Ligands: KBD, EDO, CL |
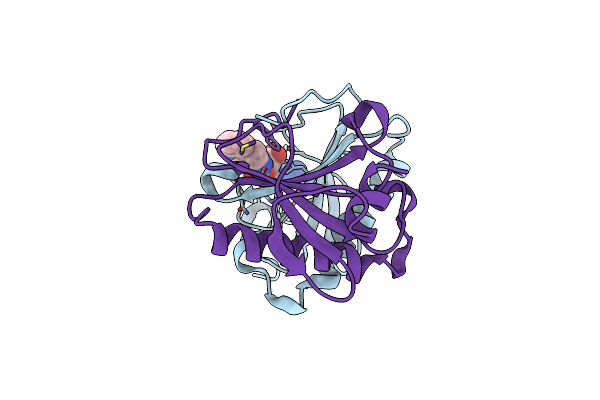 |
Human Histidine Triad Nucleotide Binding Protein 1 (Hint1) With Bound 5'-O-[(3-Indolyl)-1-Ethyl]Carbamoyl Guanosine
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2019-09-18 Classification: HYDROLASE Ligands: HHJ, CL |
 |
Organism: Enterococcus casseliflavus
Method: X-RAY DIFFRACTION Resolution:1.53 Å Release Date: 2018-02-28 Classification: TRANSFERASE/ANTIBIOTIC Ligands: CL, EDS |
 |
Crystal Structure Of The Intrinsic Colistin Resistance Enzyme Icr(Mc) From Moraxella Catarrhalis, Catalytic Domain, Thr315Ala Mutant Di-Zinc And Peg Complex
Organism: Moraxella sp. hmsc061h09
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2018-01-31 Classification: TRANSFERASE Ligands: ZN, 15P, CL |
 |
Crystal Structure Of The Intrinsic Colistin Resistance Enzyme Icr(Mc) From Moraxella Catarrhalis, Catalytic Domain, Thr315Ala Mutant Mono-Zinc And Phosphoethanolamine Complex
Organism: Moraxella sp. hmsc061h09
Method: X-RAY DIFFRACTION Resolution:1.66 Å Release Date: 2018-01-31 Classification: TRANSFERASE Ligands: ZN, OPE, SO4, 15P |
 |
Crystal Structure Of The Intrinsic Colistin Resistance Enzyme Icr(Mc) From Moraxella Catarrhalis, Catalytic Domain, Phosphate-Bound Complex
Organism: Moraxella sp. hmsc061h09
Method: X-RAY DIFFRACTION Resolution:2.61 Å Release Date: 2018-01-31 Classification: TRANSFERASE Ligands: SO4, GOL, PO4, ACT |
 |
Crystal Structure Of The Intrinsic Colistin Resistance Enzyme Icr(Mc) From Moraxella Catarrhalis, Catalytic Domain, Mono-Zinc Complex
Organism: Moraxella sp. hmsc061h09
Method: X-RAY DIFFRACTION Resolution:2.33 Å Release Date: 2018-01-31 Classification: TRANSFERASE Ligands: PO4, ZN, SO4, ACT, GOL |
 |
Crystal Structure Of Rifampin Monooxygenase From Streptomyces Venezuelae, Complexed With Rifampin And Fad
Organism: Streptomyces venezuelae (strain atcc 10712 / cbs 650.69 / dsm 40230 / jcm 4526 / nbrc 13096 / pd 04745)
Method: X-RAY DIFFRACTION Resolution:3.32 Å Release Date: 2017-12-13 Classification: OXIDOREDUCTASE Ligands: FAD, RFP, CL, MG |
 |
Crystal Structure Of Rifampin Monooxygenase From Streptomyces Venezuelae, Complex With Fad
Organism: Streptomyces venezuelae (strain atcc 10712 / cbs 650.69 / dsm 40230 / jcm 4526 / nbrc 13096 / pd 04745)
Method: X-RAY DIFFRACTION Resolution:3.39 Å Release Date: 2017-08-16 Classification: OXIDOREDUCTASE Ligands: FAD, CL, GOL |
 |
Crystal Structure Of Aminoglycoside Acetyltransferase Aac(2')-Ia In Complex With N2'-Acetylgentamicin C1A And Coenzyme A
Organism: Providencia stuartii
Method: X-RAY DIFFRACTION Resolution:2.48 Å Release Date: 2017-03-15 Classification: TRANSFERASE Ligands: 8MM, TAR, ACO, GOL, COA |
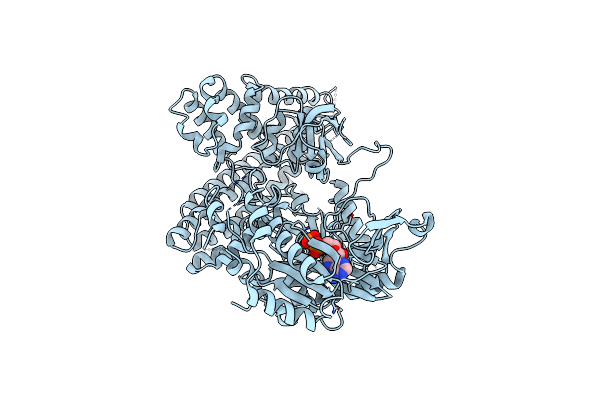 |
Crystal Structure Of Rifampin Phosphotransferase Rph-Lm From Listeria Monocytogenes In Complex With Adp And Magnesium
Organism: Listeria monocytogenes serotype 4b str. f2365
Method: X-RAY DIFFRACTION Resolution:2.59 Å Release Date: 2016-01-13 Classification: TRANSFERASE Ligands: MG, ADP |
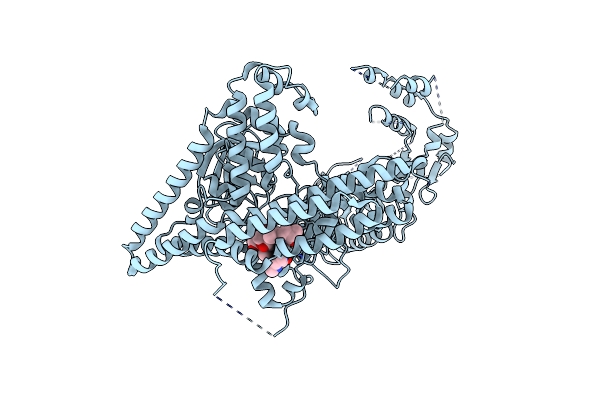 |
Crystal Structure Of Rifampin Phosphotransferase Rph-Lm From Listeria Monocytogenes In Complex With Rifampin
Organism: Listeria monocytogenes serotype 4b str. f2365
Method: X-RAY DIFFRACTION Resolution:2.70 Å Release Date: 2015-12-30 Classification: TRANSFERASE/ANTIBIOTIC Ligands: 5WQ, CL |
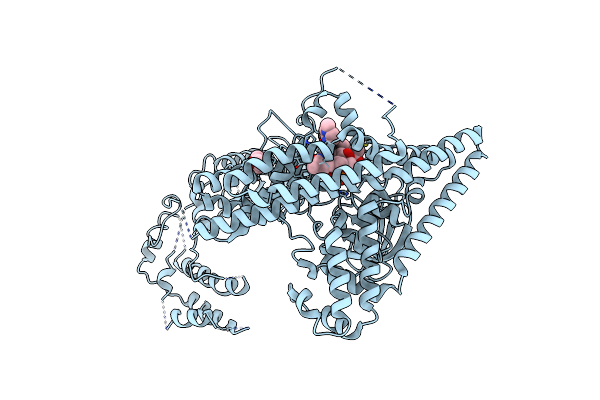 |
Crystal Structure Of Rifampin Phosphotransferase Rph-Lm From Listeria Monocytogenes In Complex With Rifampin-Phosphate
Organism: Listeria monocytogenes serotype 4b str. f2365
Method: X-RAY DIFFRACTION Resolution:2.85 Å Release Date: 2015-12-30 Classification: TRANSFERASE/ANTIBIOTIC Ligands: 5WP, CL, MPD |

