Search Count: 15
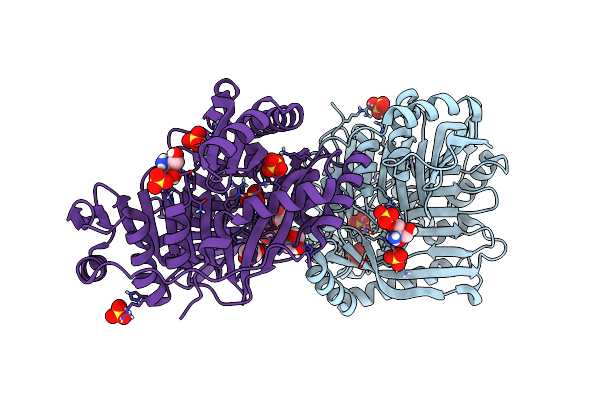 |
Crystallographic Structure Of A Salicylate Synthase From M. Abscessus (Mab-Sas)
Organism: Mycobacteroides abscessus
Method: X-RAY DIFFRACTION Resolution:2.07 Å Release Date: 2024-01-31 Classification: BIOSYNTHETIC PROTEIN Ligands: TRS, GOL, SO4, PEG |
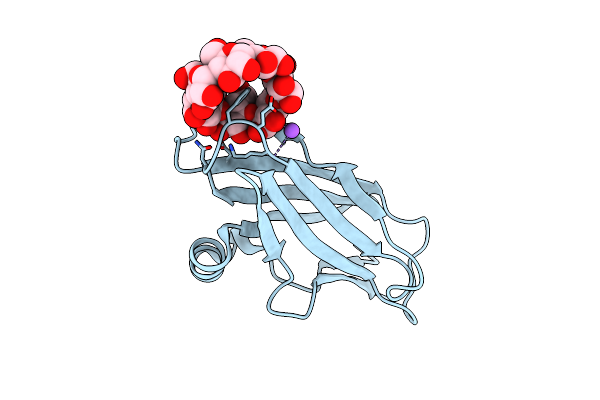 |
Orthorhombic Crystal Structure Of Ptg Cbm21 In Complex With Beta-Cyclodextrin
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.47 Å Release Date: 2022-11-02 Classification: SUGAR BINDING PROTEIN |
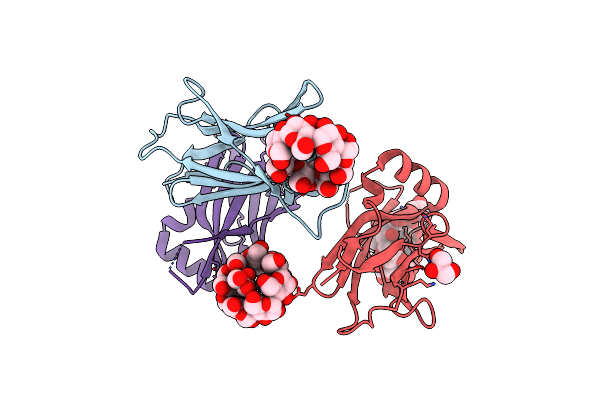 |
Monoclinic Crystal Structure Of Ptg Cbm21 In Complex With Beta-Cyclodextrin
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2022-11-02 Classification: SUGAR BINDING PROTEIN Ligands: GOL |
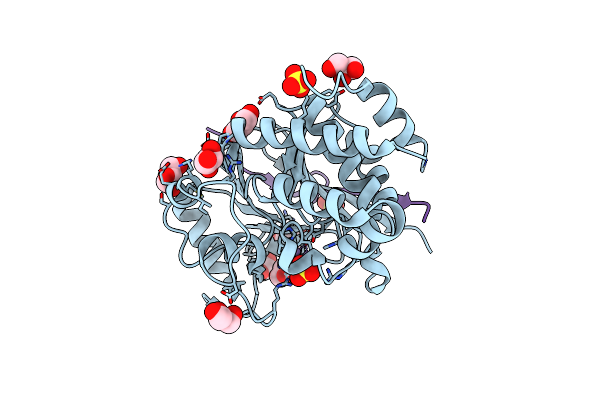 |
Crystal Structure Of Protein Phosphatase 1 In Complex With Pp1-Binding Peptide From Ptg
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.05 Å Release Date: 2022-11-02 Classification: HYDROLASE Ligands: GOL, MN, SO4 |
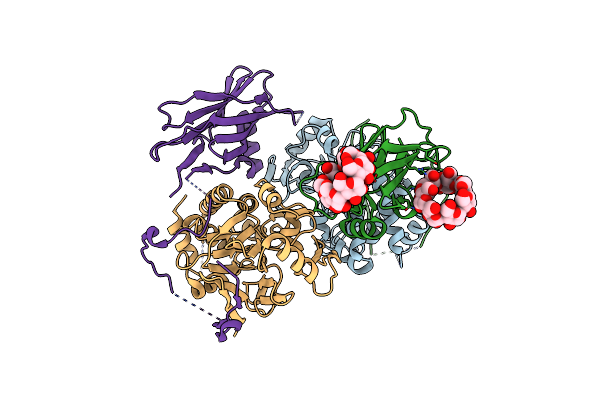 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.69 Å Release Date: 2022-11-02 Classification: HYDROLASE |
 |
Nmr Solution Structure Of Unbound Recombinant Human Nerve Growth Factor (Rhngf)
|
 |
Crystal Structure Of Human Serum Albumin In Complex With Myristic Acid At 2.27 Angstrom Resolution
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.27 Å Release Date: 2021-02-24 Classification: PROTEIN TRANSPORT Ligands: MYR, MPD, FMT, PO4 |
 |
Crystal Structure Of Human Serum Albumin In Complex With Perfluorooctanoic Acid (Pfoa) At 2.10 Angstrom Resolution
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2021-02-24 Classification: PROTEIN TRANSPORT Ligands: 8PF, MPO, PGE, PG4, MYR, MPD, EDO |
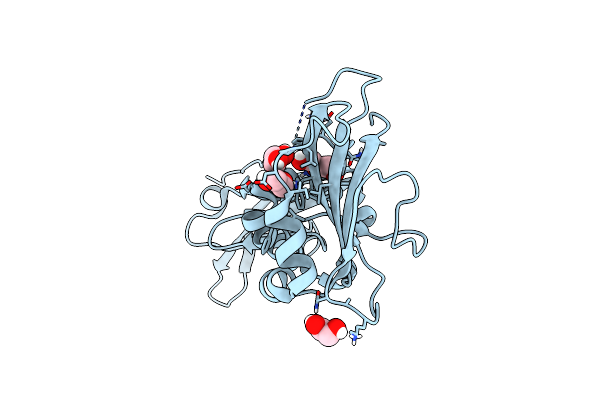 |
Crystal Structure Of The V72I Mutant Of The Mouse Alpha-Dystroglycan N-Terminal Region
Organism: Mus musculus
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2017-11-01 Classification: CELL ADHESION Ligands: EDO, PEG |
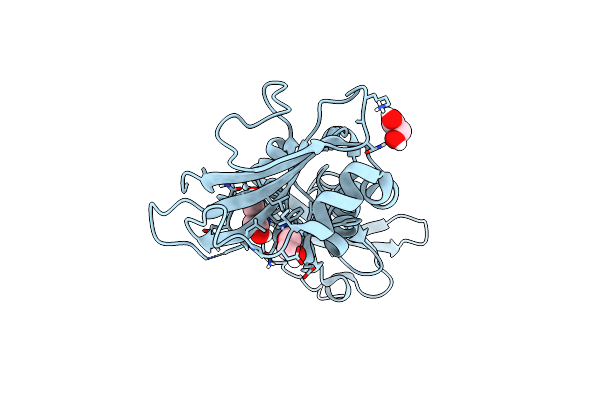 |
Crystal Structure Of The D109N Mutant Of The Mouse Alpha-Dystroglycan N-Terminal Region
Organism: Mus musculus
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2017-11-01 Classification: CELL ADHESION Ligands: EDO, PEG |
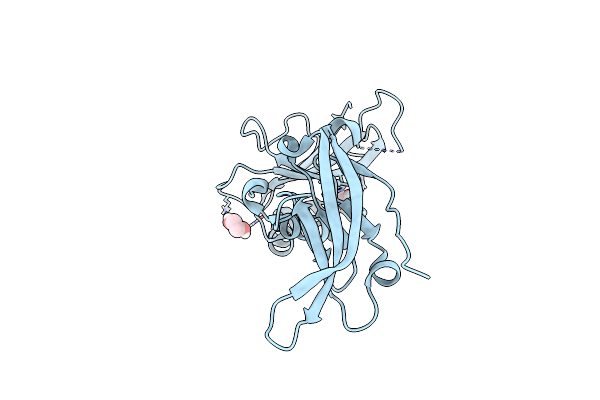 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2017-07-12 Classification: CELL ADHESION Ligands: EDO |
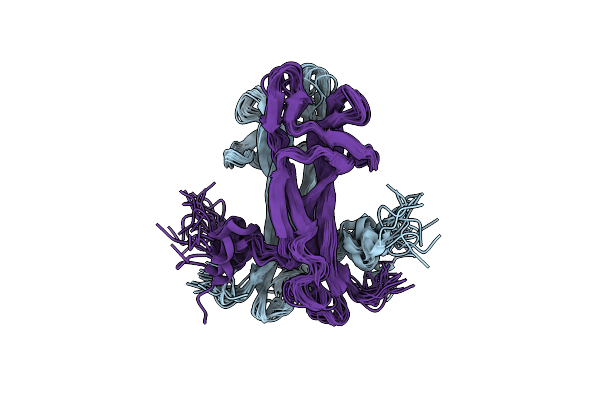 |
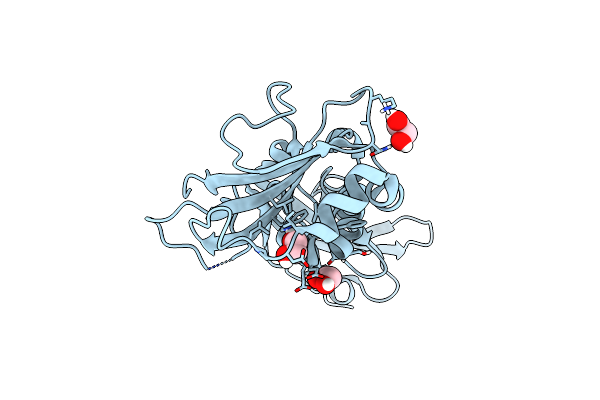 |
Organism: Mus musculus
Method: X-RAY DIFFRACTION Resolution:1.59 Å Release Date: 2015-05-13 Classification: STRUCTURAL PROTEIN Ligands: EDO, CL |
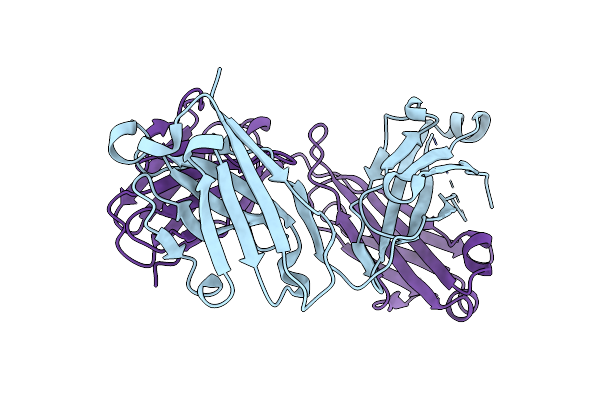 |
Organism: Rattus norvegicus
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2006-04-04 Classification: IMMUNE SYSTEM Ligands: CL |
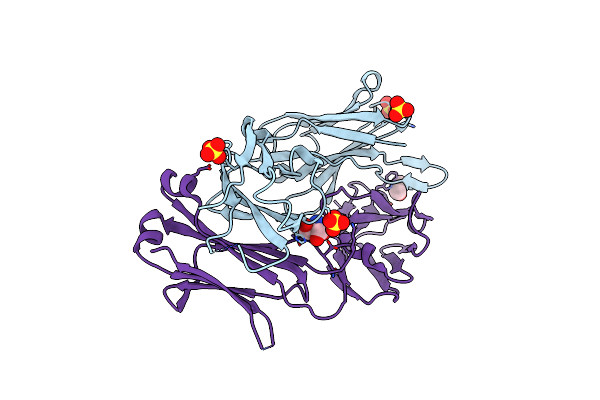 |
Organism: Mus musculus
Method: X-RAY DIFFRACTION Resolution:1.78 Å Release Date: 2005-03-01 Classification: IMMUNE SYSTEM Ligands: TRS, IPA, SO4 |

