Search Count: 28
 |
Organism: Pseudomonas aeruginosa
Method: X-RAY DIFFRACTION Resolution:1.40 Å Release Date: 2023-06-14 Classification: HYDROLASE Ligands: MES, IOD |
 |
Crystal Structure Of Choe In Complex With Acetate And Thiocholine (Crystal Form 2)
Organism: Pseudomonas aeruginosa
Method: X-RAY DIFFRACTION Resolution:1.36 Å Release Date: 2023-06-14 Classification: HYDROLASE Ligands: ACT, ETM, GOL, CL, P6G |
 |
Organism: Pseudomonas aeruginosa
Method: X-RAY DIFFRACTION Resolution:1.54 Å Release Date: 2023-06-14 Classification: HYDROLASE Ligands: AT3, GOL, CL, ETM |
 |
Crystal Structure Of Choe N147A Mutant In Complex With Thiocholine And Chloride
Organism: Pseudomonas aeruginosa
Method: X-RAY DIFFRACTION Resolution:1.38 Å Release Date: 2023-06-14 Classification: HYDROLASE Ligands: CL, ETM, GOL |
 |
Organism: Pseudomonas aeruginosa
Method: X-RAY DIFFRACTION Resolution:1.82 Å Release Date: 2023-06-14 Classification: HYDROLASE Ligands: GOL, BR |
 |
Crystal Structure Of Choe In Complex With Acetate And Tetraethylammonium (Tea)
Organism: Pseudomonas aeruginosa
Method: X-RAY DIFFRACTION Resolution:1.49 Å Release Date: 2023-06-14 Classification: HYDROLASE Ligands: ACT, NET |
 |
Crystal Structure Of Choe, A Bacterial Acetylcholinesterase From Pseudomonas Aeruginosa
Organism: Pseudomonas aeruginosa (strain atcc 15692 / dsm 22644 / cip 104116 / jcm 14847 / lmg 12228 / 1c / prs 101 / pao1)
Method: X-RAY DIFFRACTION Resolution:1.35 Å Release Date: 2020-05-13 Classification: HYDROLASE Ligands: MES, GOL, BUA, CL, P6G |
 |
Organism: Pseudomonas aeruginosa (strain atcc 15692 / dsm 22644 / cip 104116 / jcm 14847 / lmg 12228 / 1c / prs 101 / pao1)
Method: X-RAY DIFFRACTION Resolution:1.65 Å Release Date: 2020-05-13 Classification: HYDROLASE Ligands: ACT, ETM, GOL |
 |
Organism: Pseudomonas aeruginosa (strain atcc 15692 / dsm 22644 / cip 104116 / jcm 14847 / lmg 12228 / 1c / prs 101 / pao1)
Method: X-RAY DIFFRACTION Resolution:1.85 Å Release Date: 2020-05-13 Classification: HYDROLASE Ligands: QFJ, IOD, PPI, ETM |
 |
Organism: Pseudomonas aeruginosa (strain atcc 15692 / dsm 22644 / cip 104116 / jcm 14847 / lmg 12228 / 1c / prs 101 / pao1)
Method: X-RAY DIFFRACTION Resolution:1.57 Å Release Date: 2020-05-13 Classification: HYDROLASE Ligands: AT3, GOL |
 |
Crystal Structure Of Choe D285N Mutant In Complex With Acetate And Thiocholine
Organism: Pseudomonas aeruginosa (strain atcc 15692 / dsm 22644 / cip 104116 / jcm 14847 / lmg 12228 / 1c / prs 101 / pao1)
Method: X-RAY DIFFRACTION Resolution:1.43 Å Release Date: 2020-05-13 Classification: HYDROLASE Ligands: ACT, ETM |
 |
Organism: Pseudomonas aeruginosa (strain atcc 15692 / dsm 22644 / cip 104116 / jcm 14847 / lmg 12228 / 1c / prs 101 / pao1)
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2020-05-13 Classification: HYDROLASE Ligands: ETM, GOL |
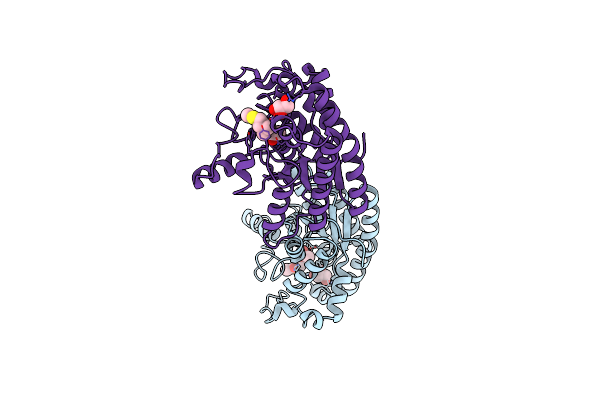 |
Crystal Structure Of Choe S38A Mutant In Complex With Acetate And Acetylthiocholine
Organism: Pseudomonas aeruginosa (strain atcc 15692 / dsm 22644 / cip 104116 / jcm 14847 / lmg 12228 / 1c / prs 101 / pao1)
Method: X-RAY DIFFRACTION Resolution:1.42 Å Release Date: 2020-05-13 Classification: HYDROLASE Ligands: ACT, AT3, GOL |
 |
Crystal Structure Of Piee, The Flavin-Dependent Monooxygenase Involved In The Biosynthesis Of Piericidin A1
Organism: Streptomyces sp. scsio 03032
Method: X-RAY DIFFRACTION Resolution:2.02 Å Release Date: 2020-03-11 Classification: FLAVOPROTEIN Ligands: FAD, PGE, 1PE, CL, GOL |
 |
Crystal Structure Of The Flavin-Dependent Monooxygenase Piee In Complex With Fad And Substrate
Organism: Streptomyces sp. scsio 03032
Method: X-RAY DIFFRACTION Resolution:2.52 Å Release Date: 2020-03-11 Classification: FLAVOPROTEIN Ligands: FAD, CL, PKS, PGE, 1PE, GOL |
 |
Crystal Structure Of Crmk, A Flavoenzyme Involved In The Shunt Product Recycling Mechanism In Caerulomycin Biosynthesis
Organism: Actinoalloteichus sp. wh1-2216-6
Method: X-RAY DIFFRACTION Resolution:1.84 Å Release Date: 2017-02-15 Classification: OXIDOREDUCTASE Ligands: FAD |
 |
Crystal Structure Of Crmk, A Flavoenzyme Involved In The Shunt Product Recycling Mechanism In Caerulomycin Biosynthesis
Organism: Actinoalloteichus sp. wh1-2216-6
Method: X-RAY DIFFRACTION Resolution:2.15 Å Release Date: 2017-02-15 Classification: OXIDOREDUCTASE Ligands: FAD, 67L, 67K |
 |
Organism: Pseudomonas putida
Method: X-RAY DIFFRACTION Resolution:1.85 Å Release Date: 2008-09-02 Classification: METAL BINDING PROTEIN Ligands: FES |
 |
Organism: Escherichia coli o157:h7
Method: X-RAY DIFFRACTION Resolution:2.05 Å Release Date: 2008-02-12 Classification: LIGAND BINDING PROTEIN, METAL TRANSPORT |
 |
Organism: Mycobacterium tuberculosis
Method: X-RAY DIFFRACTION Resolution:1.93 Å Release Date: 2007-11-06 Classification: TRANSPORT PROTEIN Ligands: CYN, SO4, HEM |

