Search Count: 34
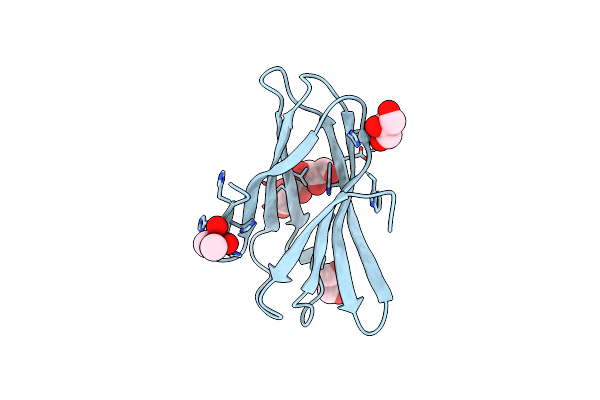 |
Structure Of A Novel Carbohydrate Binding Module From Ruminococcus Flavefaciens Fd-1 Endoglucanase Cel5A Solved At The As Edge
Organism: Ruminococcus flavefaciens
Method: X-RAY DIFFRACTION Resolution:1.29 Å Release Date: 2016-06-29 Classification: SUGAR BINDING PROTEIN Ligands: CAC, GOL |
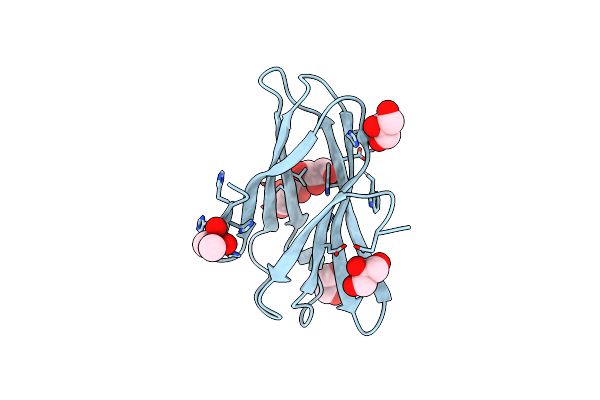 |
Very High Resolution Structure Of A Novel Carbohydrate Binding Module From Ruminococcus Flavefaciens Fd-1 Endoglucanase Cel5A
Organism: Ruminococcus flavefaciens
Method: X-RAY DIFFRACTION Resolution:1.02 Å Release Date: 2016-06-22 Classification: SUGAR BINDING PROTEN Ligands: CAC, GOL |
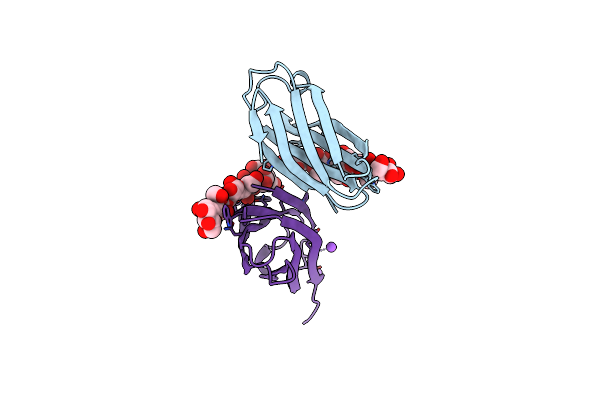 |
The Complexity Of The Ruminococcus Flavefaciens Cellulosome Reflects An Expansion In Glycan Recognition
Organism: Ruminococcus flavefaciens
Method: X-RAY DIFFRACTION Resolution:1.40 Å Release Date: 2016-06-22 Classification: SUGAR BINDING PROTEIN Ligands: CA, NA |
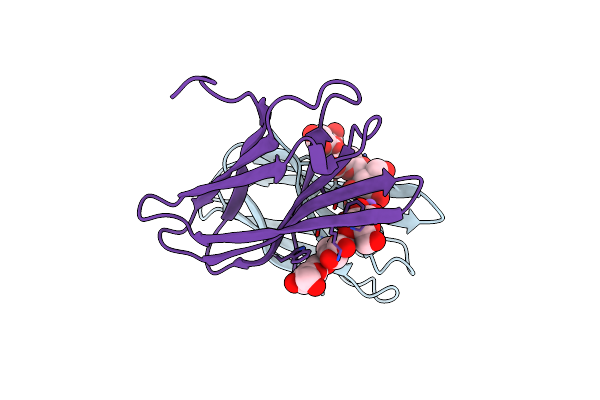 |
The Complexity Of The Ruminococcus Flavefaciens Cellulosome Reflects An Expansion In Glycan Recognition
Organism: Ruminococcus flavefaciens
Method: X-RAY DIFFRACTION Resolution:1.61 Å Release Date: 2016-06-22 Classification: SUGAR BINDING PROTEIN |
 |
The Complexity Of The Ruminococcus Flavefaciens Cellulosome Reflects An Expansion In Glycan Recognition
Organism: Ruminococcus flavefaciens
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2016-06-22 Classification: SUGAR BINDING PROTEIN |
 |
The Complexity Of The Ruminococcus Flavefaciens Cellulosome Reflects An Expansion In Glycan Recognition
Organism: Ruminococcus flavefaciens
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2016-06-22 Classification: SUGAR BINDING PROTEIN |
 |
Semet Structure Of A Novel Carbohydrate Binding Module From Glycoside Hydrolase Family 9 (Cel9A) From Ruminococcus Flavefaciens Fd-1 In The Orthorhombic Form
Organism: Ruminococcus flavefaciens
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2016-01-20 Classification: SUGAR BINDING PROTEIN Ligands: 2PE, P6G, EDO, GOL, PEG, CA, PG4, HHD |
 |
Structure Of A Novel Carbohydrate Binding Module From Glycoside Hydrolase Family 5 Glucanase From Ruminococcus Flavefaciens Fd-1
Organism: Ruminococcus flavefaciens
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2016-01-20 Classification: SUGAR BINDING PROTEIN |
 |
Semet Structure Of A Novel Carbohydrate Binding Module From Glycoside Hydrolase Family 5 Glucanase From Ruminococcus Flavefaciens Fd-1
Organism: Ruminococcus flavefaciens
Method: X-RAY DIFFRACTION Resolution:2.28 Å Release Date: 2016-01-20 Classification: SUGAR BINDING PROTEIN |
 |
Structure Of A Novel Carbohydrate Binding Module From Glycoside Hydrolase Family 5 Glucanase From Ruminococcus Flavefaciens Fd-1 Collected At The Zn Edge
Organism: Ruminococcus flavefaciens
Method: X-RAY DIFFRACTION Resolution:2.69 Å Release Date: 2016-01-20 Classification: SUGAR BINDING PROTEIN |
 |
Structure Of A Novel Carbohydrate Binding Module From Glycoside Hydrolase Family 5 Glucanase From Ruminococcus Flavefaciens Fd-1 At Medium Resolution
Organism: Ruminococcus flavefaciens
Method: X-RAY DIFFRACTION Resolution:2.59 Å Release Date: 2016-01-20 Classification: SUGAR BINDING PROTEIN |
 |
Semet Structure Of A Novel Carbohydrate Binding Module From Glycoside Hydrolase Family 9 (Cel9A) From Ruminococcus Flavefaciens Fd-1
Organism: Ruminococcus flavefaciens
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2016-01-20 Classification: SUGAR BINDING PROTEIN Ligands: 2PE, P6G, CA, BGQ |
 |
High Resolution Structure Of A Novel Carbohydrate Binding Module From Glycoside Hydrolase Family 9 (Cel9A) From Ruminococcus Flavefaciens Fd-1
Organism: Ruminococcus flavefaciens
Method: X-RAY DIFFRACTION Resolution:1.75 Å Release Date: 2016-01-20 Classification: SUGAR BINDING PROTEIN Ligands: P6G, GOL, PGE, PG4 |
 |
Crystal Structure Of A Novel Unsaturated Beta-Glucuronyl Hydrolase Enzyme, Belonging To Family Gh105, Involved In Ulvan Degradation
Organism: Nonlabens ulvanivorans
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2014-01-22 Classification: HYDROLASE Ligands: NO3 |
 |
Crystal Structure Of The Alpha-L-Arabinofuranosidase Umabf62A From Ustilago Maidys
Organism: Ustilago maydis
Method: X-RAY DIFFRACTION Resolution:1.00 Å Release Date: 2014-01-15 Classification: HYDROLASE Ligands: CA, GOL, TRS, 1PE |
 |
Crystal Structure Of The Alpha-L-Arabinofuranosidase Umabf62A From Ustilago Maydis In Complex With L-Arabinofuranose
Organism: Ustilago maydis
Method: X-RAY DIFFRACTION Resolution:1.20 Å Release Date: 2014-01-15 Classification: HYDROLASE Ligands: CA, FUB, AHR, TRS |
 |
Crystal Structure Of The Alpha-L-Arabinofuranosidase Paabf62A From Podospora Anserina In Complex With Cellotriose
Organism: Podospora anserina
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2014-01-15 Classification: HYDROLASE Ligands: CA, TRS, 1PE, EPE |
 |
Crystal Structure Of The Alpha-L-Arabinofuranosidase Paabf62A From Podospora Anserina
Organism: Podospora anserina
Method: X-RAY DIFFRACTION Resolution:1.44 Å Release Date: 2014-01-15 Classification: HYDROLASE Ligands: CA, EPE, TRS, 1PE |
 |
Organism: Bacillus subtilis
Method: X-RAY DIFFRACTION Resolution:1.65 Å Release Date: 2012-12-12 Classification: HYDROLASE Ligands: FRU, NA |
 |
Organism: Bacillus subtilis
Method: X-RAY DIFFRACTION Resolution:1.10 Å Release Date: 2012-12-12 Classification: HYDROLASE |

