Search Count: 16
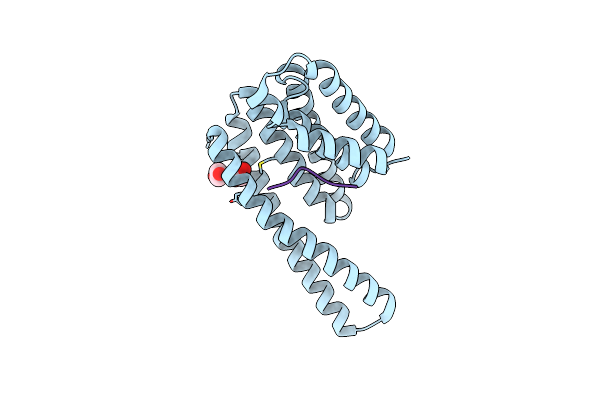 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:3.20 Å Release Date: 2023-06-07 Classification: SIGNALING PROTEIN Ligands: EDO |
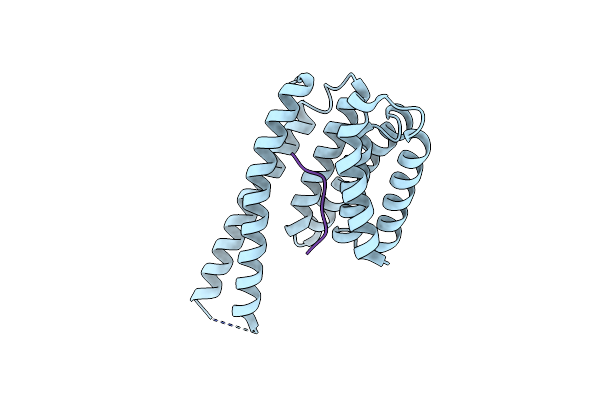 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:3.16 Å Release Date: 2023-06-07 Classification: SIGNALING PROTEIN |
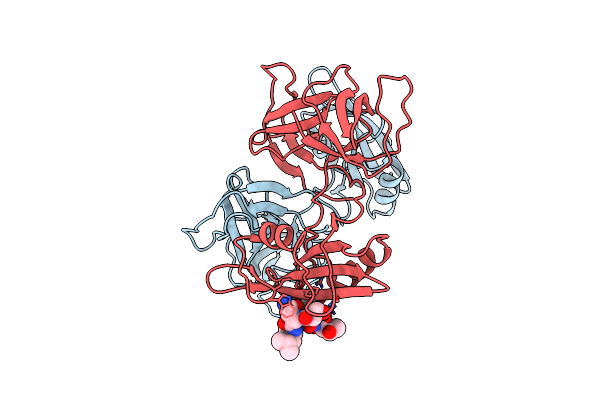 |
Growth Factor Receptor-Bound Protein 2 (Grb2) Bound To Phosphorylated Peak3 (Py24) Peptide
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2023-06-07 Classification: SIGNALING PROTEIN |
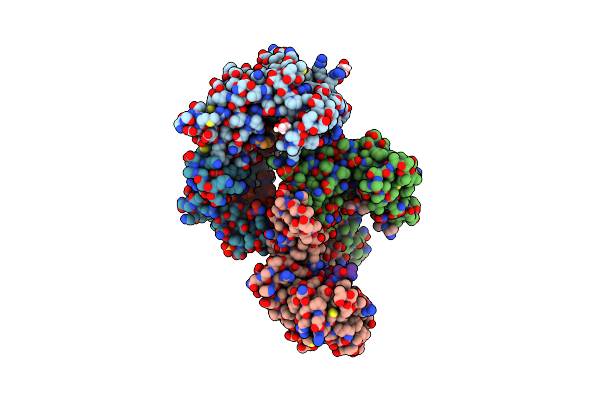 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.70 Å Release Date: 2023-06-07 Classification: SIGNALING PROTEIN Ligands: EDO, SO4 |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:3.08 Å Release Date: 2023-01-18 Classification: LIGASE Ligands: ZN |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:3.03 Å Release Date: 2023-01-18 Classification: LIGASE Ligands: ZN, SO4 |
 |
Structure Of Dimeric Phosphorylated Pediculus Humanus (Ph) Pink1 With Kinked Alpha-C Helix In Chain B
Organism: Pediculus humanus corporis
Method: ELECTRON MICROSCOPY Release Date: 2022-01-12 Classification: TRANSFERASE |
 |
Structure Of Dimeric Phosphorylated Pediculus Humanus (Ph) Pink1 With Extended Alpha-C Helix In Chain B
Organism: Pediculus humanus corporis
Method: ELECTRON MICROSCOPY Release Date: 2022-01-12 Classification: TRANSFERASE |
 |
Structure Of Dodecameric Unphosphorylated Pediculus Humanus (Ph) Pink1 D357A Mutant
Organism: Pediculus humanus corporis
Method: ELECTRON MICROSCOPY Release Date: 2022-01-12 Classification: TRANSFERASE |
 |
Structure Of Dimeric Unphosphorylated Pediculus Humanus (Ph) Pink1 D357A Mutant
Organism: Pediculus humanus corporis
Method: ELECTRON MICROSCOPY Release Date: 2022-01-12 Classification: TRANSFERASE |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.39 Å Release Date: 2022-01-05 Classification: LIGASE Ligands: ZN, GOL |
 |
Crystal Structure Of Rbr E3 Ligase Rnf216 In Complex With K63-Linked Di-Ubiquitin
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.52 Å Release Date: 2022-01-05 Classification: LIGASE Ligands: ZN, SO4, GOL |
 |
Crystal Structure Of Phosphorylated Rbr E3 Ligase Rnf216 In Complex With K63-Linked Di-Ubiquitin
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.21 Å Release Date: 2022-01-05 Classification: LIGASE Ligands: ZN, SO4, GOL, PG0 |
 |
Organism: Pediculus humanus corporis
Method: X-RAY DIFFRACTION Resolution:3.53 Å Release Date: 2021-12-22 Classification: TRANSFERASE |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.58 Å Release Date: 2018-01-17 Classification: SIGNALING PROTEIN Ligands: CL, EDO |
 |
Crystal Structure Of Human Pro-Myostatin Precursor At 4.2 A Resolution With Experimental Phases From Semet Labelling
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:4.19 Å Release Date: 2018-01-17 Classification: SIGNALING PROTEIN |

