Search Count: 22
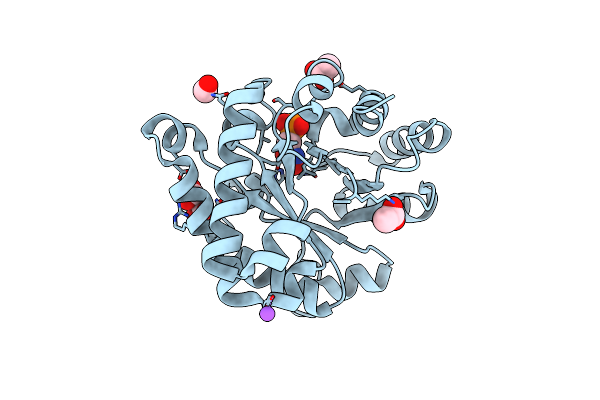 |
Crystal Structure Of The Reconstruction Of The Ancestral Triosephosphate Isomerase Of The Last Opisthokont Common Ancestor Obtained By Bayesian Inference With Pgh
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Resolution:1.61 Å Release Date: 2024-09-04 Classification: ISOMERASE |
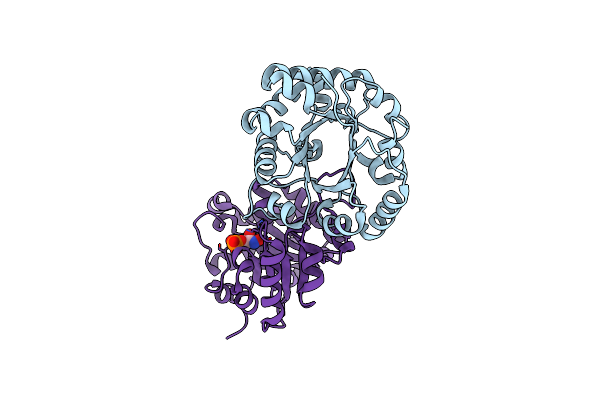 |
Crystal Structure Of The Worst Case Of The Reconstruction Of The Ancestral Triosephosphate Isomerase Of The Last Opisthokont Common Ancestor Obtained By Maximum Likelihood With Pgh
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.05 Å Release Date: 2024-09-04 Classification: ISOMERASE Ligands: PGH |
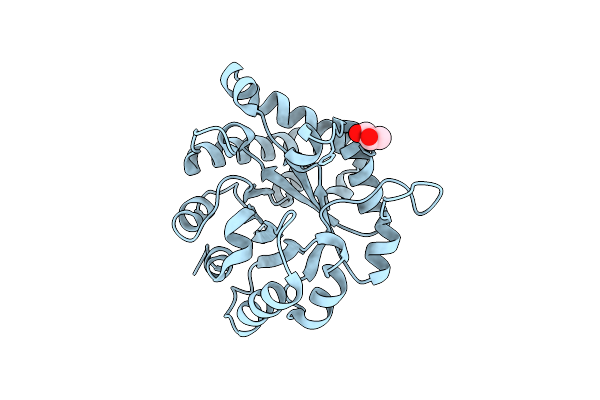 |
Crystal Structure Of The Ancestral Triosephosphate Isomerase Reconstruction Of The Last Opisthokont Common Ancestor Obtained By Bayesian Inference
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Resolution:1.38 Å Release Date: 2024-09-04 Classification: ISOMERASE Ligands: GOL |
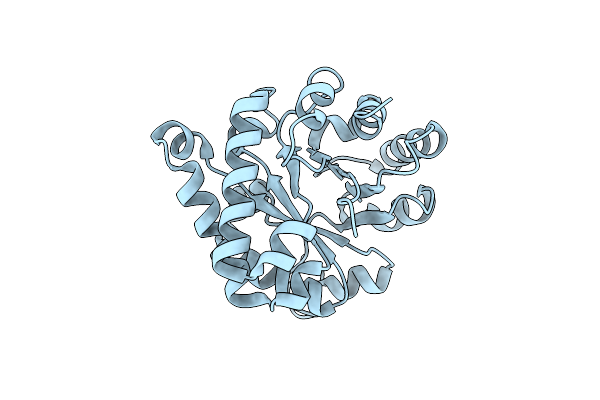 |
Crystal Structure Of The Reconstruction Of The Worst Case Of The Ancestral Triosephosphate Isomerase Of The Last Opisthokont Common Ancestor Obtained By Bayesian Inference
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Resolution:1.43 Å Release Date: 2024-09-04 Classification: ISOMERASE |
 |
Crystal Structure Of The Reconstruction Of The Ancestral Triosephosphate Isomerase Of The Last Opisthokont Common Ancestor Obtained By Maximum Likelihood
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.16 Å Release Date: 2024-09-04 Classification: ISOMERASE |
 |
Crystal Structure Of The Reconstruction Of The Ancestral Triosephosphate Isomerase Of The Last Opisthokont Common Ancestor Obtained By Maximum Likelihood With Pgh
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.06 Å Release Date: 2024-09-04 Classification: ISOMERASE Ligands: PGH, ACY, PGE, ACT, NA, EDO, PEG, CL |
 |
Crystal Structure Of The Worst Case Reconstruction Of The Ancestral Triosephosphate Isomerase Of The Last Opisthokont Common Ancestor Obtained By Maximum Likelihood
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Resolution:1.88 Å Release Date: 2024-09-04 Classification: ISOMERASE Ligands: F |
 |
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Resolution:1.64 Å Release Date: 2021-07-21 Classification: DE NOVO PROTEIN Ligands: GOL, CL |
 |
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Resolution:1.88 Å Release Date: 2021-07-21 Classification: DE NOVO PROTEIN |
 |
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.90 Å Release Date: 2021-07-21 Classification: DE NOVO PROTEIN |
 |
Crystal Structure Of Domain A From The Periplasmic Lysine-, Arginine-, Ornithine-Binding Protein (Lao) Of Salmonella Typhimurium
Organism: Salmonella typhimurium
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2021-07-14 Classification: PROTEIN BINDING Ligands: ACT, PEG |
 |
Crystal Structure Of A Reconstructed Ancestor Of Triosephosphate Isomerase From Eukaryotes
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2019-01-09 Classification: ISOMERASE Ligands: PGH |
 |
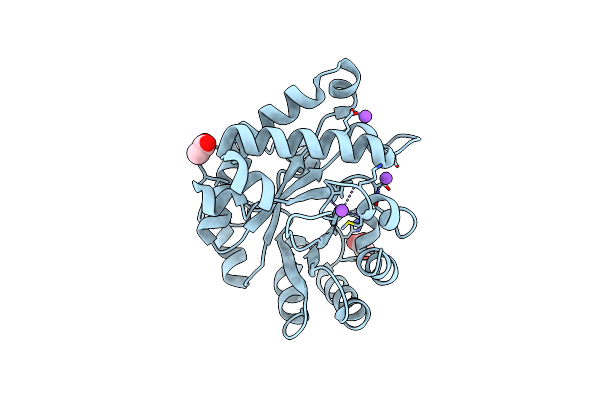
Crystal Structure Of Triosephosphate Isomerase From Zea Mays (Mexican Corn)
Organism: Zea mays
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2018-10-10 Classification: ISOMERASE Ligands: GOL, ACT |
 |
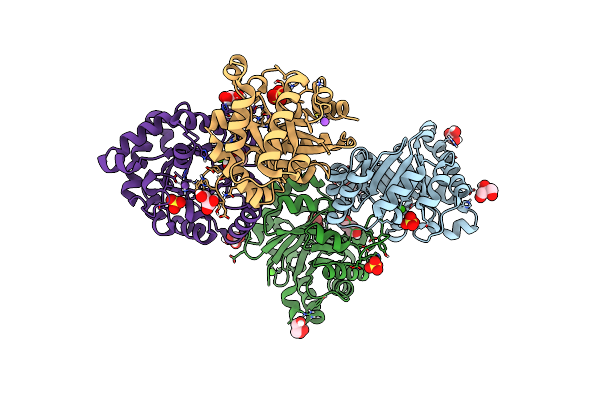
Crystal Structure Of Triosephosphate Isomerase From Clostridium Perfringens
Organism: Clostridium perfringens
Method: X-RAY DIFFRACTION Resolution:1.54 Å Release Date: 2015-07-15 Classification: ISOMERASE Ligands: ACT, NA |
 |
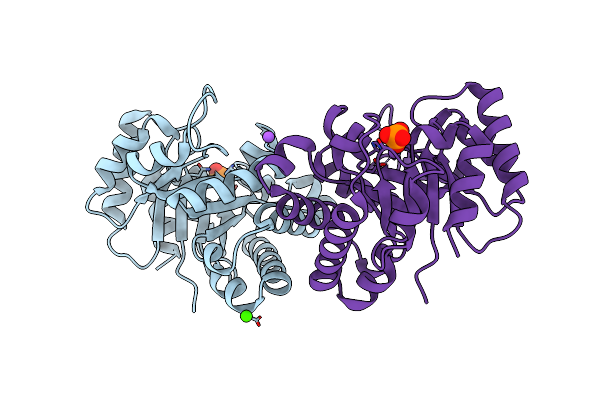
Crystal Structure Of Triosephosphate Isomerase From Deinococcus Radiodurans
Organism: Deinococcus radiodurans
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2015-07-15 Classification: ISOMERASE Ligands: SO4, GOL, CA, NA |
 |
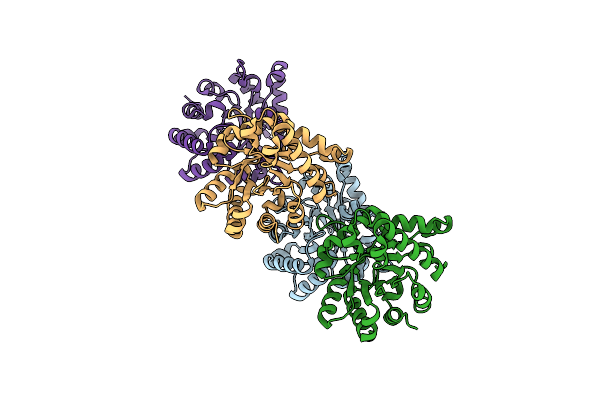
Organism: Gemmata obscuriglobus
Method: X-RAY DIFFRACTION Resolution:1.58 Å Release Date: 2015-07-15 Classification: ISOMERASE |
 |
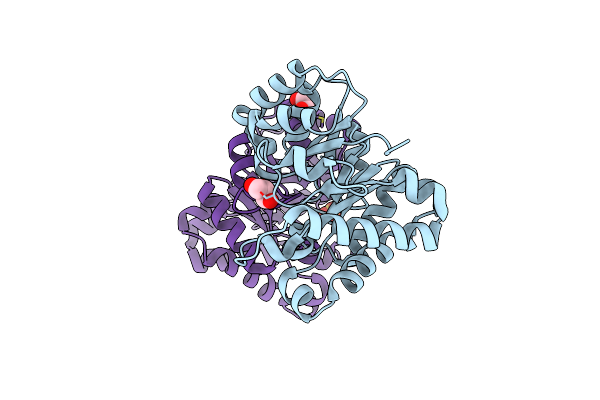
Crystal Structure Of Triosephosphate Isomerase From Streptomyces Coelicolor
Organism: Streptomyces coelicolor
Method: X-RAY DIFFRACTION Resolution:2.29 Å Release Date: 2015-07-15 Classification: ISOMERASE |
 |
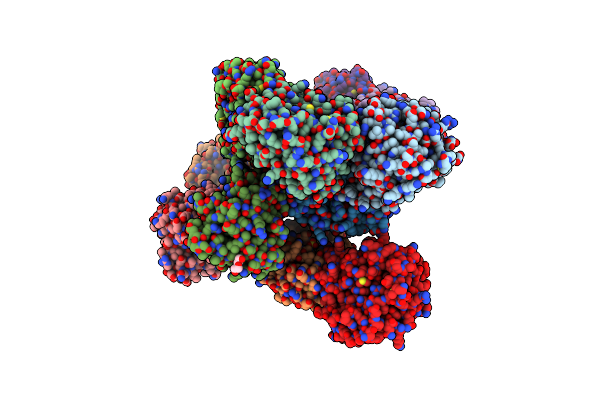
Crystal Structure Of Triosephosphate Isomerase From Trypanosoma Cruzi, Mutant E105D
Organism: Trypanosoma cruzi
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2013-10-16 Classification: ISOMERASE Ligands: GOL, SO4 |
 |
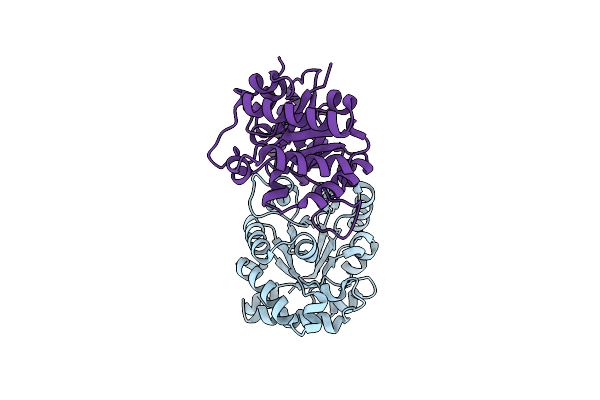
Different Contribution Of Conserved Amino Acids To The Global Properties Of Homologous Enzymes
Organism: Trypanosoma cruzi
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2013-10-02 Classification: ISOMERASE Ligands: SO4, PEG |
 |
Structural Basis Of Human Triosephosphate Isomerase Deficiency. Crystal Structure Of The Wild Type Enzyme.
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2008-07-01 Classification: ISOMERASE |

