Search Count: 49
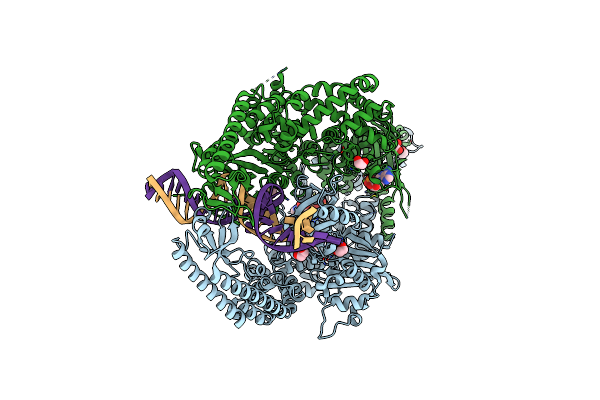 |
The Crystal Structure Of Dna-Bound Human Mutsbeta (Msh2/Msh3) In The Canonical Mismatch Bound Conformation With Adp Bound In Msh2 And Msh3
Organism: Homo sapiens, Synthetic construct
Method: X-RAY DIFFRACTION Release Date: 2025-06-11 Classification: DNA BINDING PROTEIN Ligands: ADP, MG, EDO |
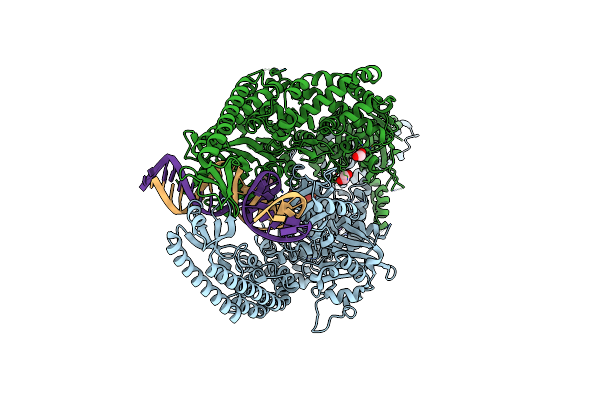 |
The Crystal Structure Of Dna-Bound Human Mutsbeta (Msh2/Msh3) In The Canonical Mismatch Bound Conformation With Adp Bound In Msh2
Organism: Homo sapiens, Synthetic construct
Method: X-RAY DIFFRACTION Release Date: 2025-06-11 Classification: DNA BINDING PROTEIN Ligands: ADP, MG, NA, EDO |
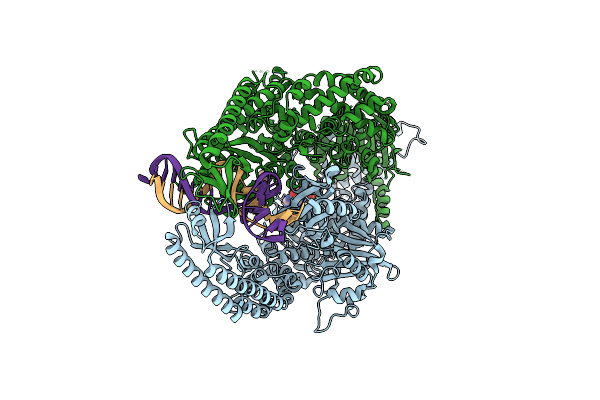 |
The Crystal Structure Of Dna-Bound Human Mutsbeta (Msh2/Msh3_G901D) In The Canonical Mismatch Bound Conformation With Adp Bound In Msh2
Organism: Homo sapiens, Synthetic construct
Method: X-RAY DIFFRACTION Release Date: 2025-06-11 Classification: DNA BINDING PROTEIN Ligands: ADP, EDO |
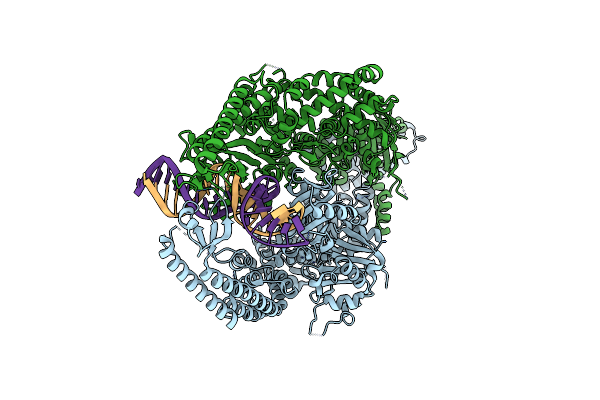 |
The Crystal Structure Of Dna-Bound Human Mutsbeta (Msh2_G674D/Msh3) In The Canonical Mismatch Bound Conformation, Nucleotide Free
Organism: Homo sapiens, Synthetic construct
Method: X-RAY DIFFRACTION Release Date: 2025-06-11 Classification: DNA BINDING PROTEIN |
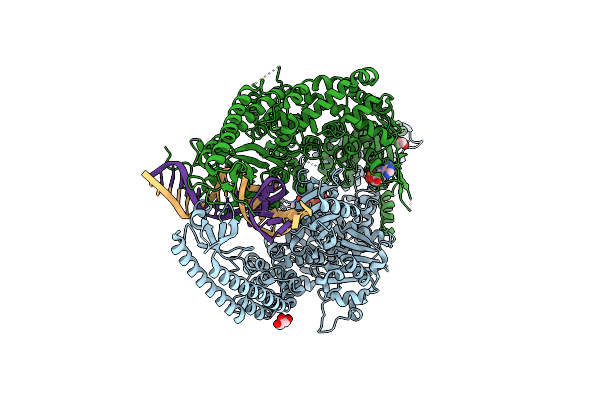 |
The Crystal Structure Of Dna-Bound Human Mutsbeta (Msh2_E749A/Msh3_E976A) In The Canonical Mismatch Bound Conformation With Adp Bound In Msh2 And Msh3
Organism: Homo sapiens, Synthetic construct
Method: X-RAY DIFFRACTION Release Date: 2025-06-11 Classification: DNA BINDING PROTEIN Ligands: ADP, GOL, CL, EDO |
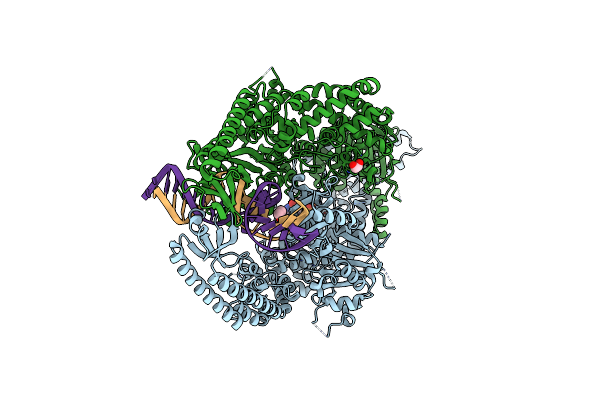 |
The Crystal Structure Of Dna-Bound Human Mutsbeta (Msh2_E749A/Msh3) In The Canonical Mismatch Bound Conformation With Adp Bound In Msh2
Organism: Homo sapiens, Synthetic construct
Method: X-RAY DIFFRACTION Release Date: 2025-06-11 Classification: DNA BINDING PROTEIN Ligands: ADP, MG, CL, EDO |
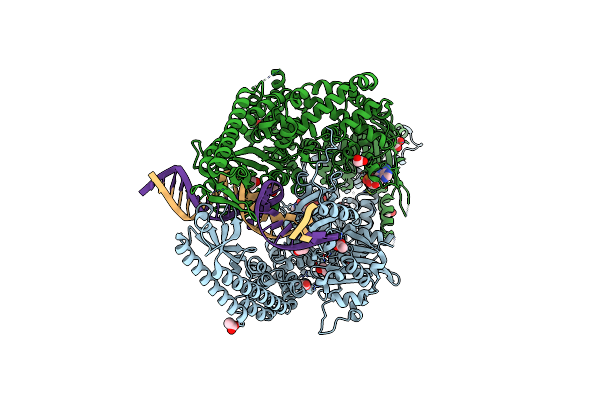 |
The Crystal Structure Of Dna-Bound Human Mutsbeta (Msh2_K675R/Msh3_K902R) In The Canonical Mismatch Bound Conformation With Adp Bound In Msh2 And Msh3
Organism: Homo sapiens, Synthetic construct
Method: X-RAY DIFFRACTION Release Date: 2025-06-11 Classification: DNA BINDING PROTEIN Ligands: ADP, MG, EDO, ACT, CL |
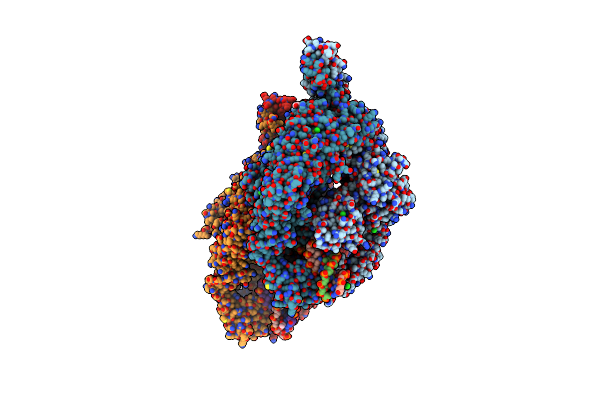 |
The Crystal Structure Of Dna-Bound Human Mutsbeta (Msh2_E749A/Msh3_E976A) In The Canonical Mismatch Bound Conformation With Adp Bound In Msh2
Organism: Homo sapiens, Synthetic construct
Method: X-RAY DIFFRACTION Release Date: 2025-06-11 Classification: DNA BINDING PROTEIN Ligands: ADP, CL, NA, PEG, EDO, GOL |
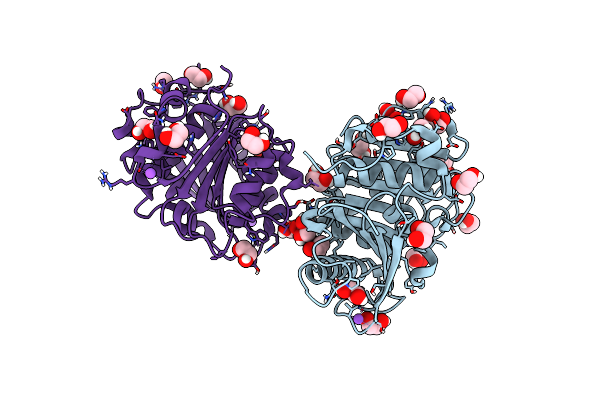 |
The Crystal Structure Of Pet44, A Petase Enzyme From Alkalilimnicola Ehrlichii
Organism: Alkalilimnicola ehrlichii
Method: X-RAY DIFFRACTION Resolution:1.40 Å Release Date: 2024-11-06 Classification: HYDROLASE |
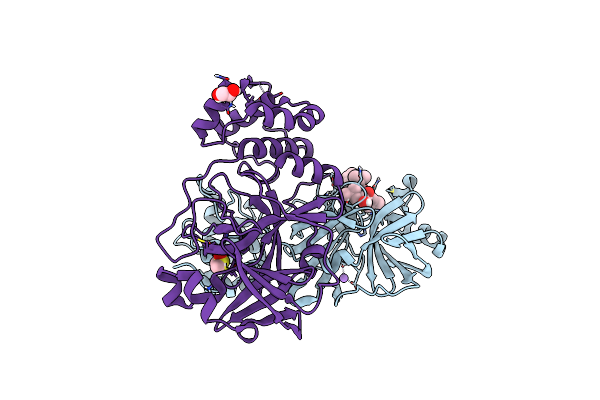 |
Crystal Structure Of The Main Protease (3Clpro/Mpro) Of Sars-Cov-2 Obtained In Presence Of 150 Micromolar Mg-132.
Organism: Severe acute respiratory syndrome coronavirus 2
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2024-05-01 Classification: VIRAL PROTEIN Ligands: ALD, CL, NA, DMS, EDO |
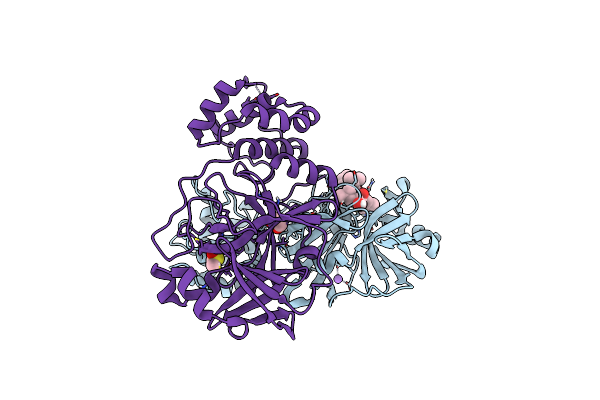 |
Crystal Structure Of The Main Protease (3Clpro/Mpro) Of Sars-Cov-2 Obtained In Presence Of 75 Micromolar Mg-132.
Organism: Severe acute respiratory syndrome coronavirus 2
Method: X-RAY DIFFRACTION Resolution:1.85 Å Release Date: 2024-05-01 Classification: VIRAL PROTEIN |
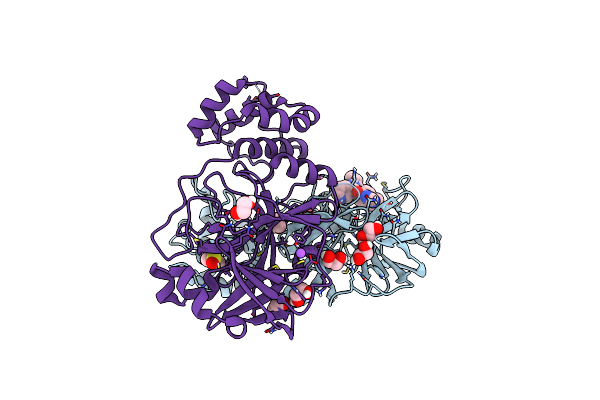 |
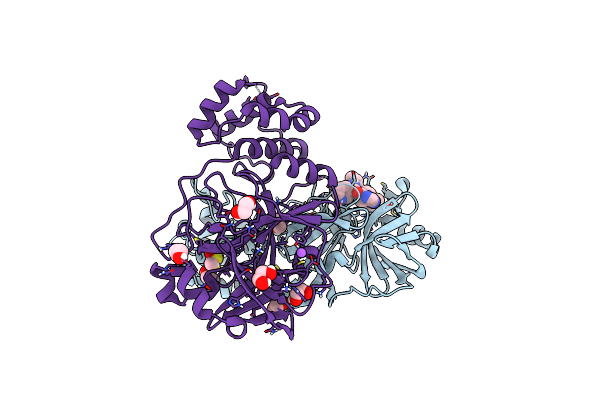
Crystal Structure Of The Main Protease (3Clpro/Mpro) Of Sars-Cov-2 Obtained In Presence Of 150 Micromolar X77.
Organism: Severe acute respiratory syndrome coronavirus 2
Method: X-RAY DIFFRACTION Resolution:1.63 Å Release Date: 2024-05-01 Classification: VIRAL PROTEIN Ligands: EDO, CL, X77, DMS, NA |
 |
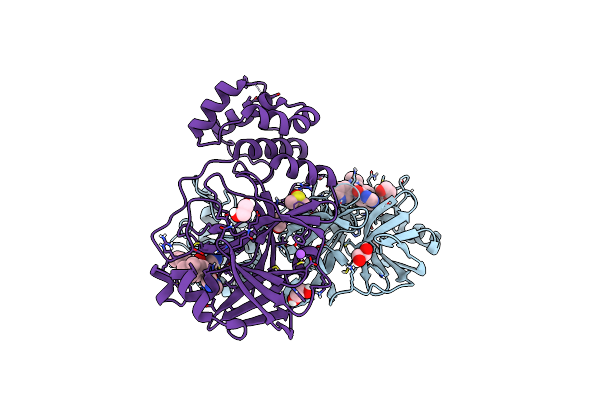
Crystal Structure Of The Main Protease (3Clpro/Mpro) Of Sars-Cov-2 Obtained In Presence Of 75 Micromolar X77.
Organism: Severe acute respiratory syndrome coronavirus 2
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2024-05-01 Classification: VIRAL PROTEIN Ligands: CL, X77, EDO, DMS, NA |
 |
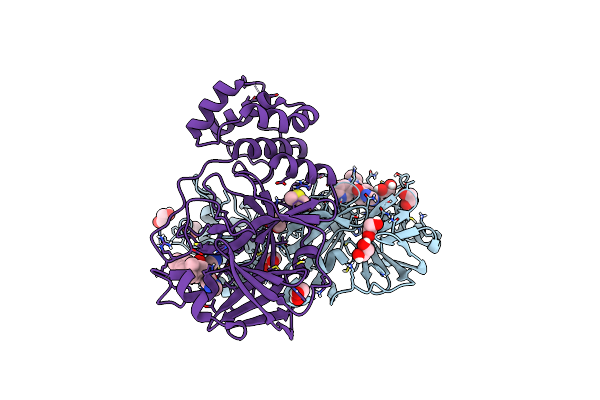
Crystal Structure Of The Main Protease (3Clpro/Mpro) Of Sars-Cov-2 Obtained In Presence Of 500 Micromolar X77 Enantiomer R.
Organism: Severe acute respiratory syndrome coronavirus 2
Method: X-RAY DIFFRACTION Resolution:1.55 Å Release Date: 2024-05-01 Classification: VIRAL PROTEIN Ligands: EDO, CL, X77, DMS, NA |
 |
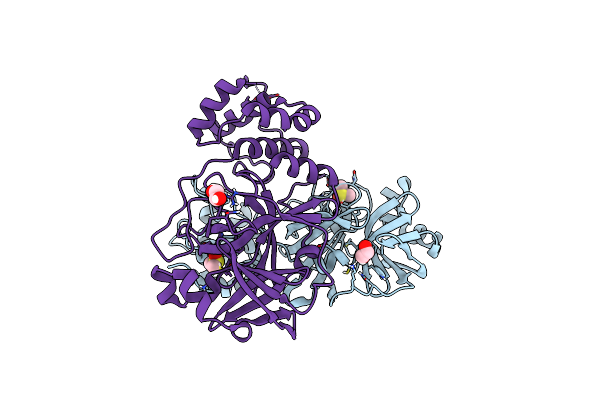
Crystal Structure Of The Main Protease (3Clpro/Mpro) Of Sars-Cov-2 Obtained In Presence Of 5 Millimolar X77 Enantiomer R.
Organism: Severe acute respiratory syndrome coronavirus 2
Method: X-RAY DIFFRACTION Resolution:1.66 Å Release Date: 2024-05-01 Classification: VIRAL PROTEIN Ligands: EDO, CL, X77, DMS, NA |
 |
Crystal Structure Of The Main Protease (3Clpro/Mpro) Of Sars-Cov-2 Obtained In Presence Of 500 Micromolar X77 Enantiomer S.
Organism: Severe acute respiratory syndrome coronavirus 2
Method: X-RAY DIFFRACTION Resolution:1.47 Å Release Date: 2024-05-01 Classification: VIRAL PROTEIN Ligands: DMS, EDO, CL, NA |
 |
Crystal Structure Of The Main Protease (3Clpro/Mpro) Of Sars-Cov-2 Obtained In Presence Of 5 Millimolar X77 Enantiomer S.
Organism: Severe acute respiratory syndrome coronavirus 2
Method: X-RAY DIFFRACTION Resolution:1.51 Å Release Date: 2024-05-01 Classification: VIRAL PROTEIN Ligands: 9M5, EDO, ACT, DMS, CL, NA |
 |
Crystal Structure Of The Main Protease (3Clpro/Mpro) Of Sars-Cov-2 Obtained In Presence Of 5 Mm Mg-132, From An "Old" Crystal.
Organism: Severe acute respiratory syndrome coronavirus 2
Method: X-RAY DIFFRACTION Resolution:1.85 Å Release Date: 2024-05-01 Classification: VIRAL PROTEIN Ligands: ALD, EDO, PEG, NA, CL |
 |
Crystal Structure Of The Main Protease (3Clpro/Mpro) Of Sars-Cov-2 Obtained In Presence Of 5 Mm X77, From An "Old" Crystal.
Organism: Severe acute respiratory syndrome coronavirus 2
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2024-05-01 Classification: VIRAL PROTEIN Ligands: ACT, EDO, CL, X77, NA |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.98 Å Release Date: 2024-01-24 Classification: TRANSFERASE Ligands: WJ9, CL |

