Search Count: 16
 |
Crystal Structure Of Methanopyrus Kandleri Malate Dehydrogenase Mutant 4 At Room Temperature
Organism: Methanopyrus kandleri
Method: X-RAY DIFFRACTION Resolution:2.39 Å Release Date: 2025-03-19 Classification: OXIDOREDUCTASE Ligands: NDP, CL, K |
 |
Organism: Methanopyrus kandleri
Method: X-RAY DIFFRACTION Resolution:1.95 Å Release Date: 2024-07-10 Classification: OXIDOREDUCTASE Ligands: NDP, CL, K |
 |
Organism: Methanopyrus kandleri
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2024-07-10 Classification: OXIDOREDUCTASE Ligands: NDP, GOL, CL, NA |
 |
Organism: Methanopyrus kandleri
Method: X-RAY DIFFRACTION Resolution:1.95 Å Release Date: 2024-07-10 Classification: OXIDOREDUCTASE Ligands: NDP, CL, NA |
 |
Organism: Selenomonas ruminantium
Method: X-RAY DIFFRACTION Resolution:3.10 Å Release Date: 2023-11-01 Classification: OXIDOREDUCTASE Ligands: PO4, NO3, CL |
 |
Crystal Structure Of Human Soluble Adenylyl Cyclase In Complex With The Inhibitor Tdi-011861
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.82 Å Release Date: 2023-03-29 Classification: SIGNALING PROTEIN Ligands: PJU |
 |
Crystal Structure Of Lactate Dehydrogenase From Selenomonas Ruminantium With Nadh.
Organism: Selenomonas ruminantium
Method: X-RAY DIFFRACTION Resolution:1.84 Å Release Date: 2021-02-10 Classification: OXIDOREDUCTASE Ligands: NAD, MLA, BO3, EDO, 1PE |
 |
Organism: Unidentified
Method: X-RAY DIFFRACTION Resolution:3.35 Å Release Date: 2014-12-24 Classification: DE NOVO PROTEIN, RNA BINDING PROTEIN |
 |
Organism: Unidentified
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2014-12-24 Classification: DE NOVO PROTEIN, RNA BINDING PROTEIN Ligands: MG |
 |
Organism: Unidentified
Method: X-RAY DIFFRACTION Resolution:2.60 Å Release Date: 2014-12-24 Classification: DE NOVO PROTEIN, RNA BINDING PROTEIN Ligands: CA |
 |
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Resolution:3.85 Å Release Date: 2014-12-24 Classification: DE NOVO PROTEIN |
 |
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Resolution:3.70 Å Release Date: 2014-12-24 Classification: DE NOVO PROTEIN |
 |
Organism: Arabidopsis thaliana
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2013-11-13 Classification: LYASE Ligands: HEZ, CO |
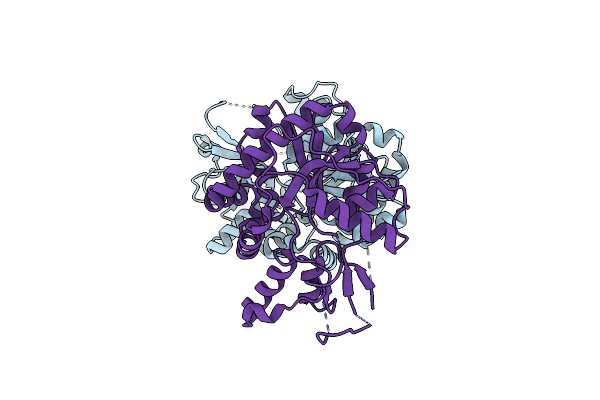 |
Crystal Structure Of Semet Derivative Of Hmp Synthase Thi5 From S. Cerevisiae
Organism: Saccharomyces cerevisiae
Method: X-RAY DIFFRACTION Resolution:2.60 Å Release Date: 2012-10-17 Classification: TRANSFERASE |
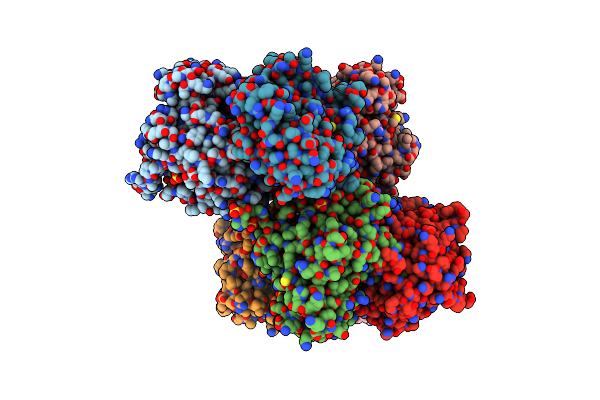 |
Organism: Saccharomyces cerevisiae
Method: X-RAY DIFFRACTION Resolution:2.70 Å Release Date: 2012-10-17 Classification: TRANSFERASE Ligands: SO4, PLP |
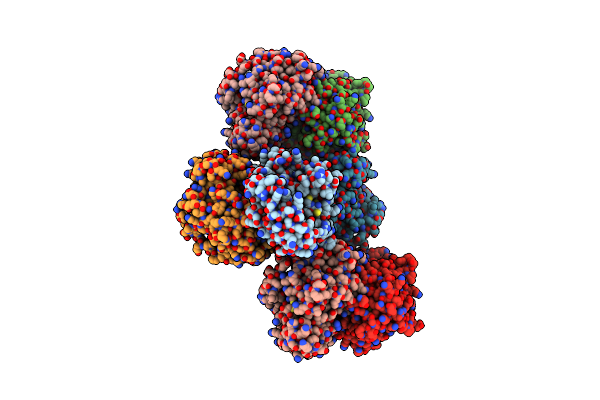 |
Organism: Saccharomyces cerevisiae
Method: X-RAY DIFFRACTION Resolution:2.90 Å Release Date: 2012-10-17 Classification: TRANSFERASE Ligands: PLP |

