Search Count: 118
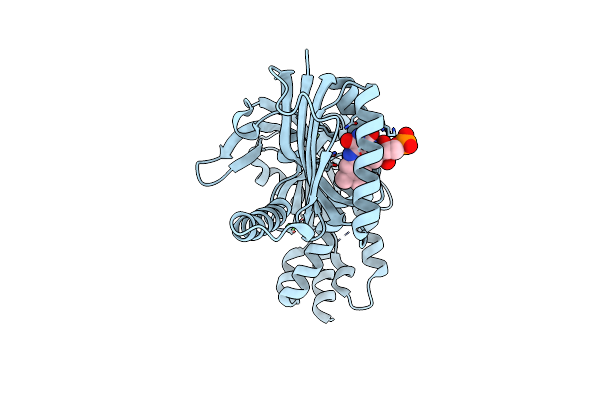 |
Sfx Room Temperature Structure Of Oscillatoria Acuminata Adenylyate Cyclase.
Organism: Oscillatoria acuminata
Method: X-RAY DIFFRACTION Resolution:1.75 Å Release Date: 2025-02-26 Classification: LYASE Ligands: FAD, CA |
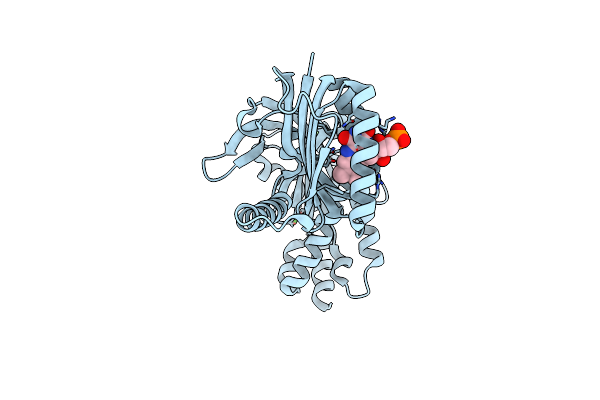 |
Ssx Room Temperature Structure Of Oscillatoria Acuminata Adenylyate Cyclase.
Organism: Oscillatoria acuminata
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2025-02-26 Classification: LYASE Ligands: FMN, CA |
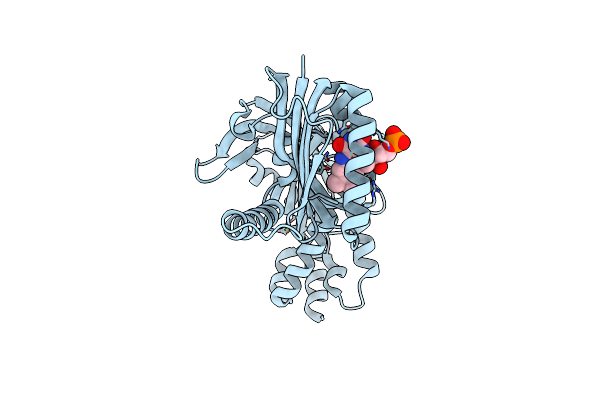 |
Organism: Oscillatoria acuminata
Method: X-RAY DIFFRACTION Resolution:1.95 Å Release Date: 2025-02-26 Classification: LYASE Ligands: FAD, CA |
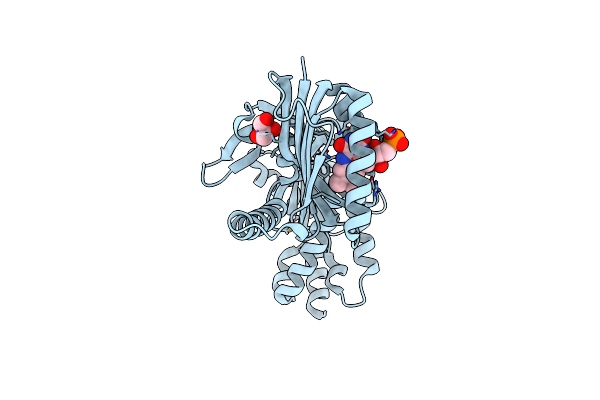 |
Organism: Oscillatoria acuminata
Method: X-RAY DIFFRACTION Resolution:1.95 Å Release Date: 2025-02-26 Classification: LYASE Ligands: FMN, GOL, CA |
 |
Serial Microseconds Crystallography At Id29 Using Sacla Extruder: Thaumatin
Organism: Thaumatococcus daniellii
Method: X-RAY DIFFRACTION Resolution:1.75 Å Release Date: 2025-01-22 Classification: PLANT PROTEIN Ligands: TLA |
 |
Organism: Thaumatococcus daniellii
Method: X-RAY DIFFRACTION Resolution:1.75 Å Release Date: 2025-01-22 Classification: PLANT PROTEIN Ligands: TLA |
 |
Organism: Thaumatococcus daniellii
Method: X-RAY DIFFRACTION Resolution:1.75 Å Release Date: 2025-01-22 Classification: PLANT PROTEIN Ligands: TLA |
 |
Serial Microseconds Crystallography At Id29 Using Fixed-Target (Small Foils): Proteinase K With 10 Um Spacing Between X-Ray Pulses
Organism: Parengyodontium album
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2025-01-22 Classification: HYDROLASE Ligands: CA, NO3 |
 |
Serial Microseconds Crystallography At Id29 Using Fixed-Target (Small Foils): Proteinase K With 20 Um Spacing Between X-Ray Pulses
Organism: Parengyodontium album
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2025-01-22 Classification: HYDROLASE Ligands: CA, NO3 |
 |
Serial Microseconds Crystallography At Id29 Using Fixed-Target (Large Foils): Proteinase K With 50 Um Spacing Between X-Ray Pulses
Organism: Parengyodontium album
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2025-01-22 Classification: HYDROLASE Ligands: CA, NO3 |
 |
Serial Microseconds Crystallography At Id29 Using Fixed-Target (Si Chip): Lysozyme - Without Ligand Glcnac (Apo)
Organism: Gallus gallus
Method: X-RAY DIFFRACTION Resolution:2.05 Å Release Date: 2025-01-22 Classification: HYDROLASE |
 |
Serial Microseconds Crystallography At Id29 Using Fixed-Target (Si Chip): Lysozyme - With Ligand Glcnac
Organism: Gallus gallus
Method: X-RAY DIFFRACTION Resolution:2.05 Å Release Date: 2025-01-22 Classification: HYDROLASE |
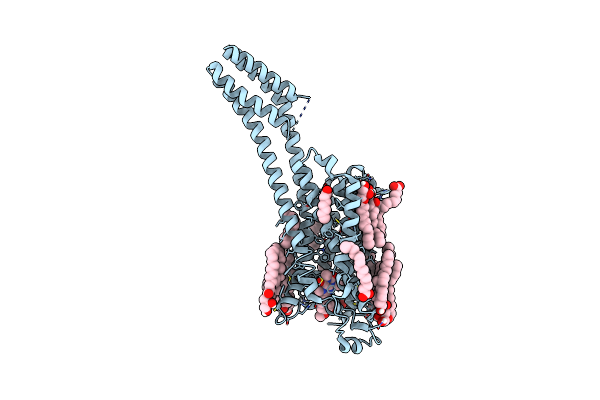 |
Serial Microseconds Crystallography At Id29 Using Fixed-Target (Small Foils): A2A Adenosine Receptor Co-Crystallised With Istradefylline
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2025-01-22 Classification: SIGNALING PROTEIN |
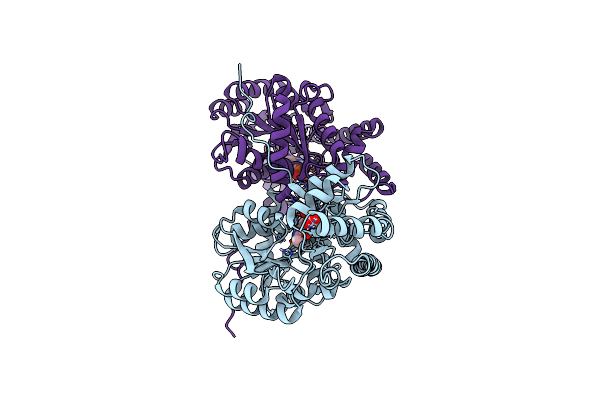 |
Joint X-Ray/Neutron Structure Of Aspastate Aminotransferase (Aat) In Complex With Pyridoxamine 5'-Phosphate (Pmp)
Organism: Sus scrofa
Method: X-RAY DIFFRACTION, NEUTRON DIFFRACTION Resolution:1.70 Å, 2.22 Å Release Date: 2022-09-28 Classification: TRANSFERASE Ligands: PLA, PMP, DOD |
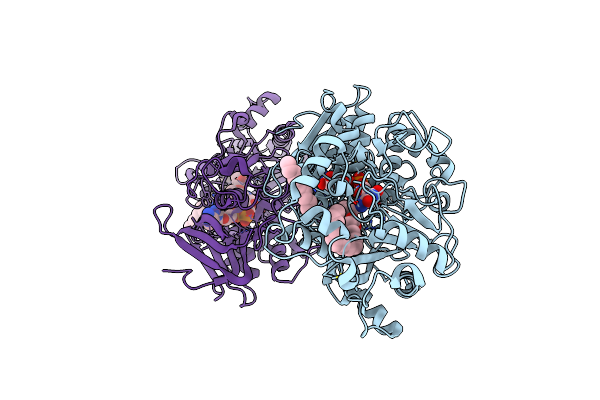 |
Difference-Refined Structure Of Fatty Acid Photodecarboxylase 20 Ps Following 400-Nm Laser Irradiation Of The Dark-State Determined By Sfx
Organism: Chlorella variabilis
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2022-09-14 Classification: OXIDOREDUCTASE Ligands: FAD, STE |
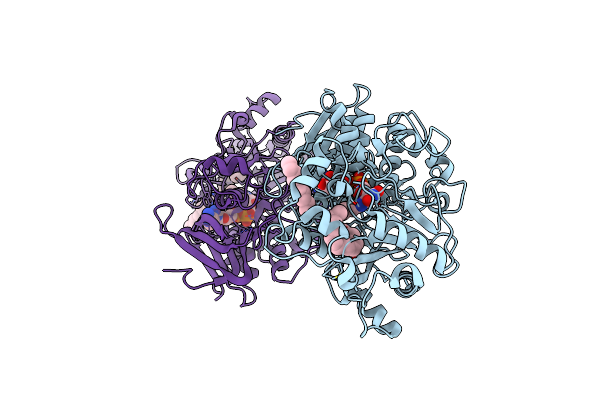 |
Difference-Refined Structure Of Fatty Acid Photodecarboxylase 900 Ps Following 400-Nm Laser Irradiation Of The Dark-State Determined By Sfx
Organism: Chlorella variabilis
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2022-09-14 Classification: OXIDOREDUCTASE Ligands: FAD, STE |
 |
Difference-Refined Structure Of Fatty Acid Photodecarboxylase 300 Ns Following 400-Nm Laser Irradiation Of The Dark-State Determined By Sfx
Organism: Chlorella variabilis
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2022-09-14 Classification: OXIDOREDUCTASE Ligands: FAD, STE |
 |
Difference-Refined Structure Of Fatty Acid Photodecarboxylase 2 Microsecond Following 400-Nm Laser Irradiation Of The Dark-State Determined By Sfx
Organism: Chlorella variabilis
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2022-09-14 Classification: OXIDOREDUCTASE Ligands: FAD, STE |
 |
Organism: Aequorea victoria
Method: X-RAY DIFFRACTION Resolution:1.63 Å Release Date: 2022-08-24 Classification: FLUORESCENT PROTEIN Ligands: GOL |
 |
Organism: Aequorea victoria
Method: X-RAY DIFFRACTION Resolution:1.63 Å Release Date: 2022-08-24 Classification: FLUORESCENT PROTEIN Ligands: GOL |

